Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T55922
(Former ID: TTDR00414)
|
|||||
| Target Name |
S-adenosylmethionine decarboxylase proenzyme (AMD1)
|
|||||
| Synonyms |
SamDC; S-adenosylmethioninedecarboxylase; AdoMetDC; AMD
Click to Show/Hide
|
|||||
| Gene Name |
AMD1
|
|||||
| Target Type |
Successful target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Acidosis [ICD-11: 5C73] | |||||
| Function |
Promotes maintenance and self-renewal of embryonic stem cells, by maintaining spermine levels. Essential for biosynthesis of the polyamines spermidine and spermine.
Click to Show/Hide
|
|||||
| BioChemical Class |
Carbon-carbon lyase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 4.1.1.50
|
|||||
| Sequence |
MEAAHFFEGTEKLLEVWFSRQQPDANQGSGDLRTIPRSEWDILLKDVQCSIISVTKTDKQ
EAYVLSESSMFVSKRRFILKTCGTTLLLKALVPLLKLARDYSGFDSIQSFFYSRKNFMKP SHQGYPHRNFQEEIEFLNAIFPNGAAYCMGRMNSDCWYLYTLDFPESRVISQPDQTLEIL MSELDPAVMDQFYMKDGVTAKDVTRESGIRDLIPGSVIDATMFNPCGYSMNGMKSDGTYW TIHITPEPEFSYVSFETNLSQTSYDDLIRKVVEVFKPGKFVTTLFVNQSSKCRTVLASPQ KIEGFKRLDCQSAMFNDYNFVFTSFAKKQQQQQS Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| ADReCS ID | BADD_A05554 | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Approved Drug(s) | [+] 1 Approved Drugs | + | ||||
| 1 | Tromethamine | Drug Info | Approved | Acidosis | [2], [3] | |
| Clinical Trial Drug(s) | [+] 1 Clinical Trial Drugs | + | ||||
| 1 | ORG 34517/34850 | Drug Info | Phase 2 | Solid tumour/cancer | [4], [5] | |
| Discontinued Drug(s) | [+] 3 Discontinued Drugs | + | ||||
| 1 | Putrescine | Drug Info | Discontinued in Phase 2 | Burn and burn infection | [6], [7] | |
| 2 | CGP-40215A | Drug Info | Terminated | Pneumocystis pneumonia | [8] | |
| 3 | MGBG | Drug Info | Terminated | Head and neck cancer | [5], [9] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Inhibitor | [+] 11 Inhibitor drugs | + | ||||
| 1 | Tromethamine | Drug Info | [1] | |||
| 2 | ORG 34517/34850 | Drug Info | [10] | |||
| 3 | Putrescine | Drug Info | [1] | |||
| 4 | MGBG | Drug Info | [10], [11] | |||
| 5 | 5'-([(Z)-4-amino-2-butenyl]methylamino)-5'-deoxyadenosine | Drug Info | [5] | |||
| 6 | 5'-Deoxy-5'-(N,N-dimethylamino)-8-methyladenosine | Drug Info | [12] | |||
| 7 | 5'-Deoxy-5'-(N,N-dimethylamino)adenosine | Drug Info | [12] | |||
| 8 | 5'-Deoxy-5'-dimethylsulfonioadenosine chloride | Drug Info | [12] | |||
| 9 | 5'-deoxy-5'-[(3-hydrazinopropyl)methylamino]adenosine | Drug Info | [5] | |||
| 10 | Hydroxyalanine | Drug Info | [13] | |||
| 11 | [(2-aminooxyethyl)methylamino]-5'-deoxyadenosine | Drug Info | [12] | |||
| Modulator | [+] 1 Modulator drugs | + | ||||
| 1 | CGP-40215A | Drug Info | [5], [8] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Pyruvic acid | Ligand Info | |||||
| Structure Description | Human AdoMetDC D174N mutant complexed with S-Adenosylmethionine methyl ester and no putrescine bound | PDB:3EP6 | ||||
| Method | X-ray diffraction | Resolution | 1.70 Å | Mutation | Yes | [14] |
| PDB Sequence |
> Chain A
SMFVSKRRFI 78 LKTCGTTLLL88 KALVPLLKLA98 RDYSGFDSIQ108 SFFYSRKNFM118 KPSHQGYPHR 128 NFQEEIEFLN138 AIFPNGAAYC148 MGRMNSDCWY158 LYTLDFNQTL177 EILMSELDPA 187 VMDQFYMKDG197 VTAKDVTRES207 GIRDLIPGSV217 IDATMFNPCG227 YSMNGMKSDG 237 TYWTIHITPE247 PEFSYVSFET257 NLSQTSYDDL267 IRKVVEVFKP277 GKFVTTLFVN 287 QPQKIEGFKR307 LDCQSAMFND317 YNFVFTSFAK327 > Chain B AHFFEGTEKL 13 LEVWFSRGSG30 DLRTIPRSEW40 DILLKDVQCS50 IISVTKTDKQ60 EAYVLSE |
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Ligand Name: Urea | Ligand Info | |||||
| Structure Description | Human AdoMetDC E178Q mutant with no putrescine bound | PDB:3EP5 | ||||
| Method | X-ray diffraction | Resolution | 1.99 Å | Mutation | Yes | [14] |
| PDB Sequence |
> Chain A
SMFVSKRRFI 78 LKTCGTTLLL88 KALVPLLKLA98 RDYSGFDSIQ108 SFFYSRKNFM118 KPSHQGYPHR 128 NFQEEIEFLN138 AIFPNGAAYC148 MGRMNSDCWY158 LYTLDFPDQT176 LQILMSELDP 186 AVMDQFYMKD196 GVTAKDVTRE206 SGIRDLIPGS216 VIDATMFNPC226 GYSMNGMKSD 236 GTYWTIHITP246 EPEFSYVSFE256 TNLSQTSYDD266 LIRKVVEVFK276 PGKFVTTLFV 286 NKIEGFKRLD309 CQSAMFNDYN319 FVFTSFAKK> Chain B HFFEGTEKLL 14 EVWFSRQGSG30 DLRTIPRSEW40 DILLKDVQCS50 IISVTKTDKQ60 EAYVLSE |
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
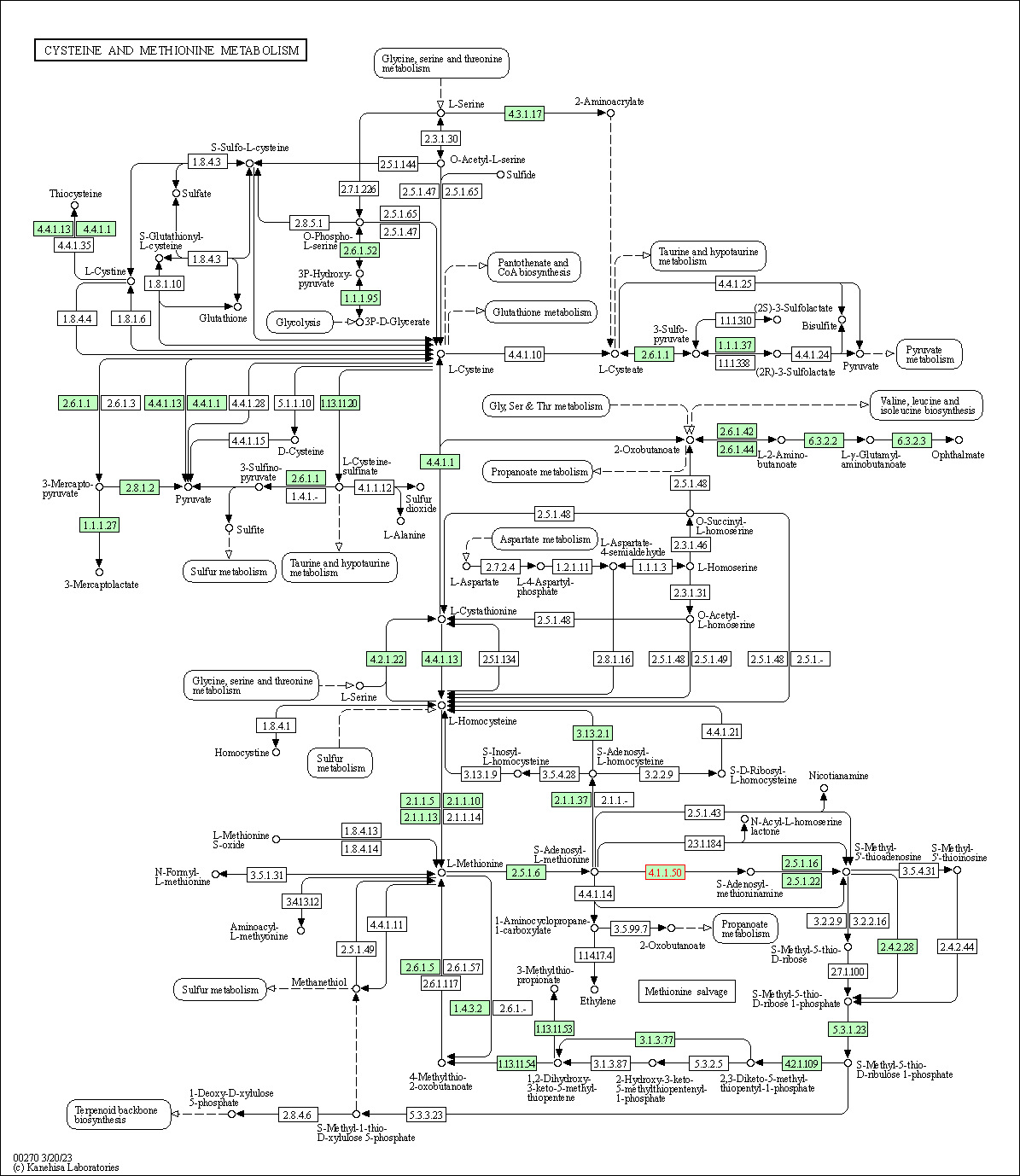
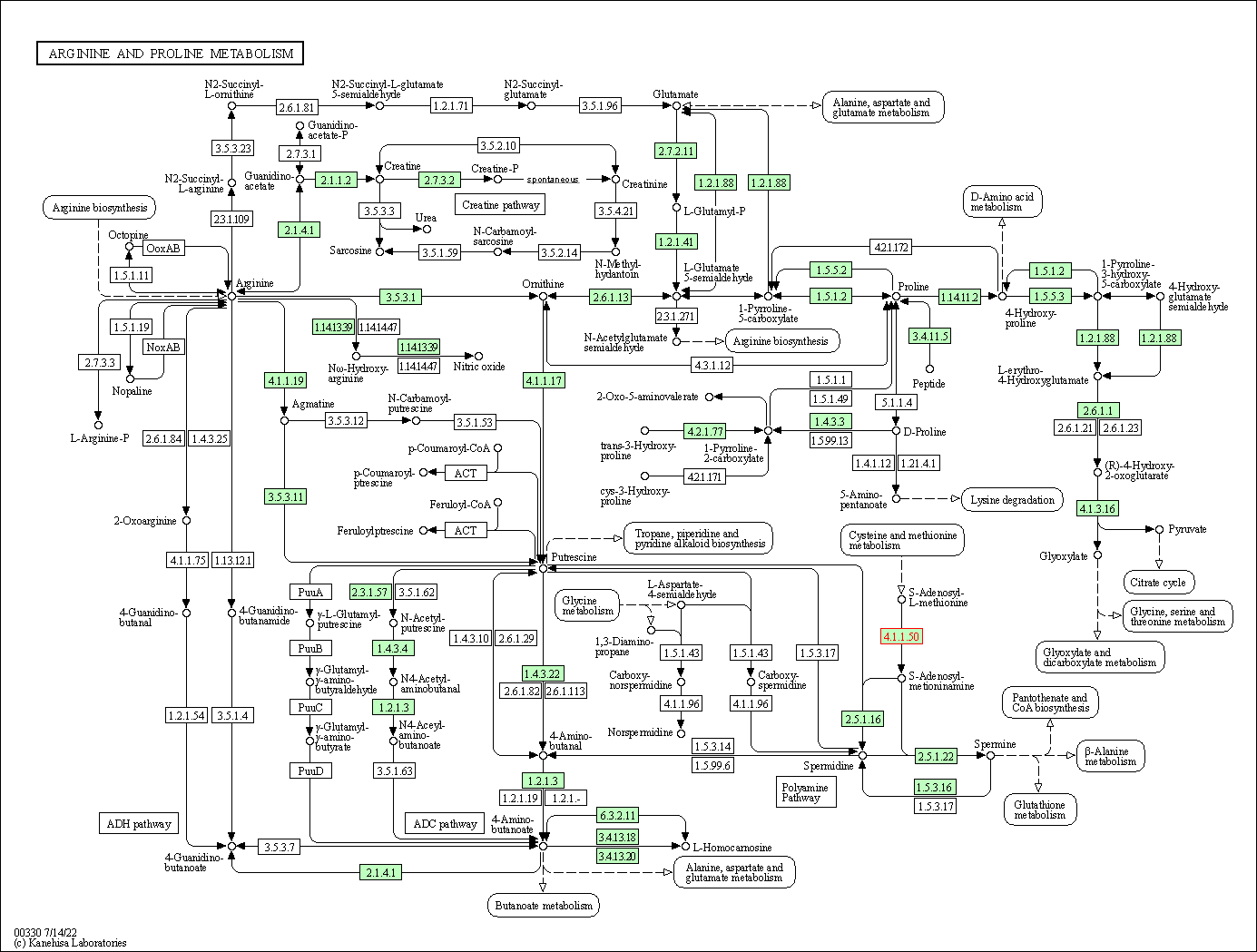
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Cysteine and methionine metabolism | hsa00270 | Affiliated Target |

|
| Class: Metabolism => Amino acid metabolism | Pathway Hierarchy | ||
| Arginine and proline metabolism | hsa00330 | Affiliated Target |

|
| Class: Metabolism => Amino acid metabolism | Pathway Hierarchy | ||
| Degree | 5 | Degree centrality | 5.37E-04 | Betweenness centrality | 2.08E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 1.51E-01 | Radiality | 1.20E+01 | Clustering coefficient | 2.00E-01 |
| Neighborhood connectivity | 6.20E+00 | Topological coefficient | 3.33E-01 | Eccentricity | 13 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) |
||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| BioCyc | [+] 3 BioCyc Pathways | + | ||||
| 1 | Methionine salvage cycle III | |||||
| 2 | Spermine biosynthesis | |||||
| 3 | Spermidine biosynthesis | |||||
| KEGG Pathway | [+] 3 KEGG Pathways | + | ||||
| 1 | Cysteine and methionine metabolism | |||||
| 2 | Arginine and proline metabolism | |||||
| 3 | Metabolic pathways | |||||
| NetPath Pathway | [+] 1 NetPath Pathways | + | ||||
| 1 | EGFR1 Signaling Pathway | |||||
| Pathwhiz Pathway | [+] 2 Pathwhiz Pathways | + | ||||
| 1 | Spermidine and Spermine Biosynthesis | |||||
| 2 | Methionine Metabolism | |||||
| WikiPathways | [+] 1 WikiPathways | + | ||||
| 1 | Metabolism of amino acids and derivatives | |||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | How many drug targets are there Nat Rev Drug Discov. 2006 Dec;5(12):993-6. | |||||
| REF 2 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7328). | |||||
| REF 3 | Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services. 2015 | |||||
| REF 4 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 5268). | |||||
| REF 5 | S-adenosylmethionine decarboxylase as an enzyme target for therapy. Pharmacol Ther. 1992 Dec;56(3):359-77. | |||||
| REF 6 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 2388). | |||||
| REF 7 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800013366) | |||||
| REF 8 | Antileishmanial effect of a potent S-adenosylmethionine decarboxylase inhibitor: CGP 40215A. Pharmacol Res. 1996 Jan;33(1):67-70. | |||||
| REF 9 | Spermine deficiency resulting from targeted disruption of the spermine synthase gene in embryonic stem cells leads to enhanced sensitivity to antiproliferative drugs. Mol Pharmacol. 2001 Feb;59(2):231-8. | |||||
| REF 10 | A phase I study of a new polyamine biosynthesis inhibitor, SAM486A, in cancer patients with solid tumours. Br J Cancer. 2000 Sep;83(5):594-601. | |||||
| REF 11 | New S-adenosylmethionine decarboxylase inhibitors with potent antitumor activity. Cancer Res. 1992 Sep 1;52(17):4712-8. | |||||
| REF 12 | New insights into the design of inhibitors of human S-adenosylmethionine decarboxylase: studies of adenine C8 substitution in structural analogues ... J Med Chem. 2009 Mar 12;52(5):1388-407. | |||||
| REF 13 | DrugBank 3.0: a comprehensive resource for 'omics' research on drugs. Nucleic Acids Res. 2011 Jan;39(Database issue):D1035-41. | |||||
| REF 14 | Structural basis for putrescine activation of human S-adenosylmethionine decarboxylase. Biochemistry. 2008 Dec 16;47(50):13404-17. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

