Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T59929
(Former ID: TTDR00251)
|
|||||
| Target Name |
Thymidine phosphorylase (TYMP)
|
|||||
| Synonyms |
TdRPase; TYMP; TP; Platelet-derived endothelial cell growth factor; PDECGF; PD-ECGF; Gliostatin
Click to Show/Hide
|
|||||
| Gene Name |
TYMP
|
|||||
| Target Type |
Successful target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Depression [ICD-11: 6A70-6A7Z] | |||||
| Function |
Catalyzes the reversible phosphorolysis of thymidine. The produced molecules are then utilized as carbon and energy sources or in the rescue of pyrimidine bases for nucleotide synthesis.
Click to Show/Hide
|
|||||
| BioChemical Class |
Pentosyltransferase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.4.2.4
|
|||||
| Sequence |
MAALMTPGTGAPPAPGDFSGEGSQGLPDPSPEPKQLPELIRMKRDGGRLSEADIRGFVAA
VVNGSAQGAQIGAMLMAIRLRGMDLEETSVLTQALAQSGQQLEWPEAWRQQLVDKHSTGG VGDKVSLVLAPALAACGCKVPMISGRGLGHTGGTLDKLESIPGFNVIQSPEQMQVLLDQA GCCIVGQSEQLVPADGILYAARDVTATVDSLPLITASILSKKLVEGLSALVVDVKFGGAA VFPNQEQARELAKTLVGVGASLGLRVAAALTAMDKPLGRCVGHALEVEEALLCMDGAGPP DLRDLVTTLGGALLWLSGHAGTQAQGAARVAAALDDGSALGRFERMLAAQGVDPGLARAL CSGSPAERRQLLPRAREQEELLAPADGTVELVRALPLALVLHELGAGRSRAGEPLRLGVG AELLVDVGQRLRRGTPWLRVHRDGPALSGPQSRALQEALVLSDRAPFAAPSPFAELVLPP Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| HIT2.0 ID | T55VJ9 | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Approved Drug(s) | [+] 1 Approved Drugs | + | ||||
| 1 | Uridine | Drug Info | Approved | Depression | [2], [3] | |
| Mode of Action | [+] 1 Modes of Action | + | ||||
| Inhibitor | [+] 24 Inhibitor drugs | + | ||||
| 1 | Uridine | Drug Info | [1] | |||
| 2 | 1-(cyclohexyl)methyl-5'-O-tritylinosine | Drug Info | [4] | |||
| 3 | 1-(cyclopropyl)methyl-5'-O-tritylinosine | Drug Info | [4] | |||
| 4 | 1-allyl-5'-O-tritylinosine | Drug Info | [4] | |||
| 5 | 1-benzyl-5'-O-tritylinosine | Drug Info | [4] | |||
| 6 | 1-propyl-5'-O-tritylinosine | Drug Info | [4] | |||
| 7 | 2'-aminoimidazolylmethyluracils | Drug Info | [5] | |||
| 8 | 5-benzyl-6-chloropyrimidine-2,4(1H,3H)-dione | Drug Info | [6] | |||
| 9 | 5-bromo-6-(cyclopropylamino)uracil hydrochloride | Drug Info | [7] | |||
| 10 | 5-bromo-6-hydrazinouracil hydrochloride | Drug Info | [7] | |||
| 11 | 5-butyl-6-chloropyrimidine-2,4(1H,3H)-dione | Drug Info | [6] | |||
| 12 | 5-chloro-6-hydrazinouracil hydrochloride | Drug Info | [7] | |||
| 13 | 5-fluoro-6-[(2-aminoimidazol-1-yl)methyl]uracil | Drug Info | [8] | |||
| 14 | 6-amino-5-bromouracil | Drug Info | [9] | |||
| 15 | 6-amino-5-chlorouracil hydrochloride | Drug Info | [7] | |||
| 16 | 6-bromo-5-phenylpyrimidine-2,4(1H,3H)-dione | Drug Info | [6] | |||
| 17 | 6-chloro-5-(2-thienyl)pyrimidine-2,4(1H,3H)-dione | Drug Info | [6] | |||
| 18 | 6-chloro-5-heptylpyrimidine-2,4(1H,3H)-dione | Drug Info | [6] | |||
| 19 | 6-chloro-5-hexylpyrimidine-2,4(1H,3H)-dione | Drug Info | [6] | |||
| 20 | 6-chloro-5-pentylpyrimidine-2,4(1H,3H)-dione | Drug Info | [6] | |||
| 21 | 6-chloro-5-phenylpyrimidine-2,4(1H,3H)-dione | Drug Info | [6] | |||
| 22 | 6-chloro-5-propylpyrimidine-2,4(1H,3H)-dione | Drug Info | [6] | |||
| 23 | 6-fluoro-5-phenylpyrimidine-2,4(1H,3H)-dione | Drug Info | [6] | |||
| 24 | Thymine | Drug Info | [10] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Thymine | Ligand Info | |||||
| Structure Description | Structural basis for non-competitive product inhibition in human thymidine phosphorylase: implication for drug design | PDB:2J0F | ||||
| Method | X-ray diffraction | Resolution | 2.31 Å | Mutation | No | [11] |
| PDB Sequence |
KQLPELIRMK
43 RDGGRLSEAD53 IRGFVAAVVN63 GSAQGAQIGA73 MLMAIRLRGM83 DLEETSVLTQ 93 ALAQSGQQLE103 WPEAWRQQLV113 DKHSTGGVGD123 KVSLVLAPAL133 AACGCKVPMI 143 SGRGLGHTGG153 TLDKLESIPG163 FNVIQSPEQM173 QVLLDQAGCC183 IVGQSEQLVP 193 ADGILYAARD203 VTATVDSLPL213 ITASILSKKL223 VEGLSALVVD233 VKFGAGAVFP 243 NQEQARELAK253 TLVGVGASLG263 LRVAAALTAM273 DKPLGRCVGH283 ALEVEEALLC 293 MDGAGPPDLR303 DLVTTLGGAL313 LWLSGHAGTQ323 AQGAARVAAA333 LDDGSALGRF 343 ERMLAAQGVD353 PGLARALCSG363 SPAERRQLLP373 RAREQEELLA383 PADGTVELVR 393 ALPLALVLHE403 LGAGRSRAGE413 PLRLGVGAEL423 LVDVGQRLRR433 GTPWLRVHRD 443 GPALSGPQSR453 ALQEALVLSD463 RAPFAAPLPF473 AELVLP
|
|||||
|
|
||||||
| Ligand Name: 5-Iodouracil | Ligand Info | |||||
| Structure Description | Structural features of native human thymidine phosphorylase and in complex with 5-iodouracil | PDB:2WK6 | ||||
| Method | X-ray diffraction | Resolution | 2.50 Å | Mutation | No | [12] |
| PDB Sequence |
QLPELIRMKR
44 DGGRLSEADI54 RGFVAAVVNG64 SAQGAQIGAM74 LMAIRLRGMD84 LEETSVLTQA 94 LAQSGQQLEW104 PEAWRQQLVD114 KHSTGGVGDK124 VSLVLAPALA134 ACGCKVPMIS 144 GRGLGHTGGT154 LDKLESIPGF164 NVIQSPEQMQ174 VLLDQAGCCI184 VGQSEQLVPA 194 DGILYAARDV204 TATVDSLPLI214 TASILSKKLV224 EGLSALVVDV234 KFGGAAVFPN 244 QEQARELAKT254 LVGVGASLGL264 RVAAALTAMD274 KPLGRCVGHA284 LEVEEALLCM 294 DGAGPPDLRD304 LVTTLGGALL314 WLSGHAGTQA324 QGAARVAAAL334 DDGSALGRFE 344 RMLAAQGVDP354 GLARALCSGS364 PAERRQLLPR374 AREQEELLAP384 ADGTVELVRA 394 LPLALVLHEL404 GAGRSRAGEP414 LRLGVGAELL424 VDVGQRLRRG434 TPWLRVHRDG 444 PALSGPQSRA454 LQEALVLSDR464 APFAAPSPFA474 ELVLPPQ
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Pyrimidine metabolism | hsa00240 | Affiliated Target |
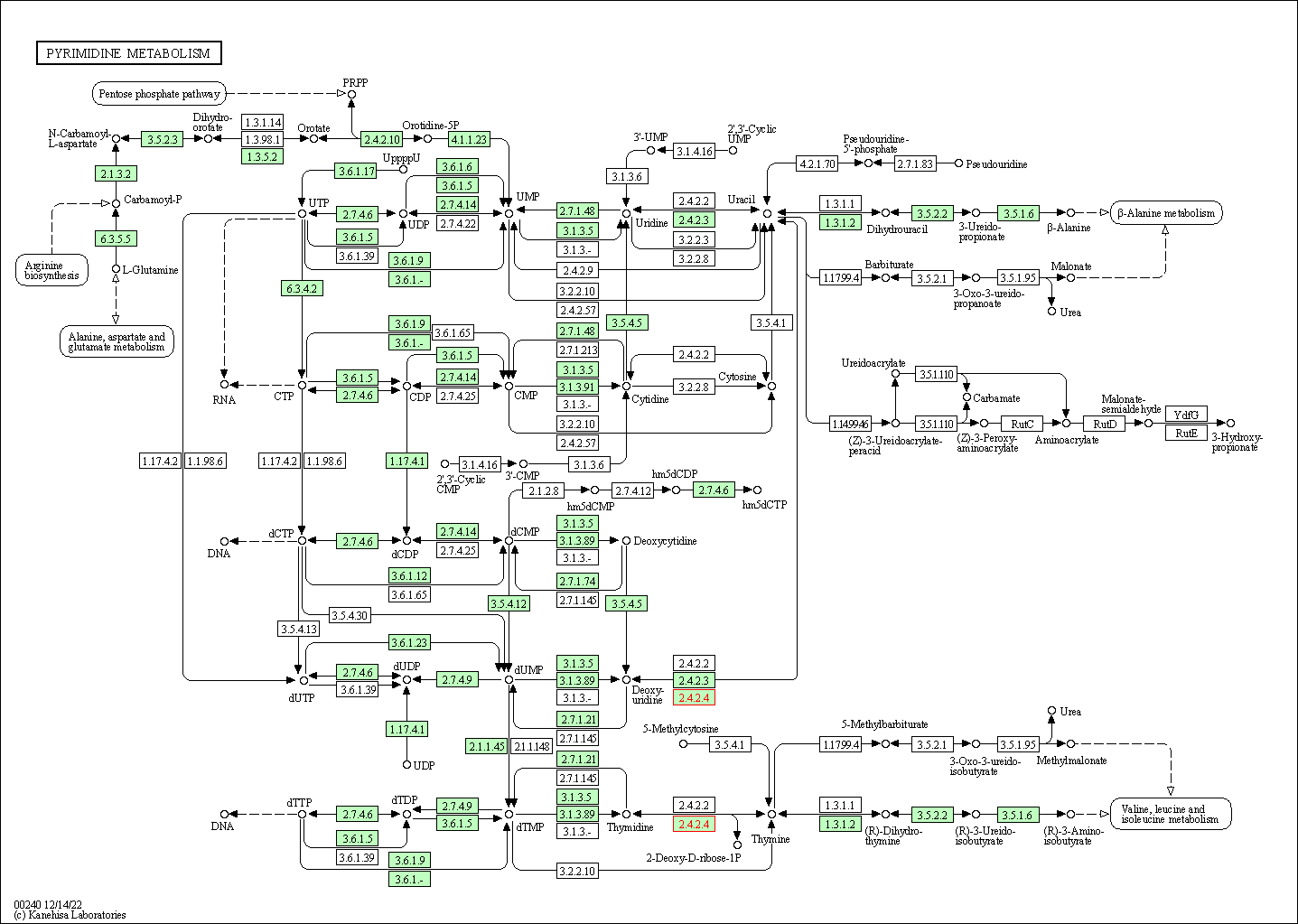
|
| Class: Metabolism => Nucleotide metabolism | Pathway Hierarchy | ||
| Drug metabolism - other enzymes | hsa00983 | Affiliated Target |
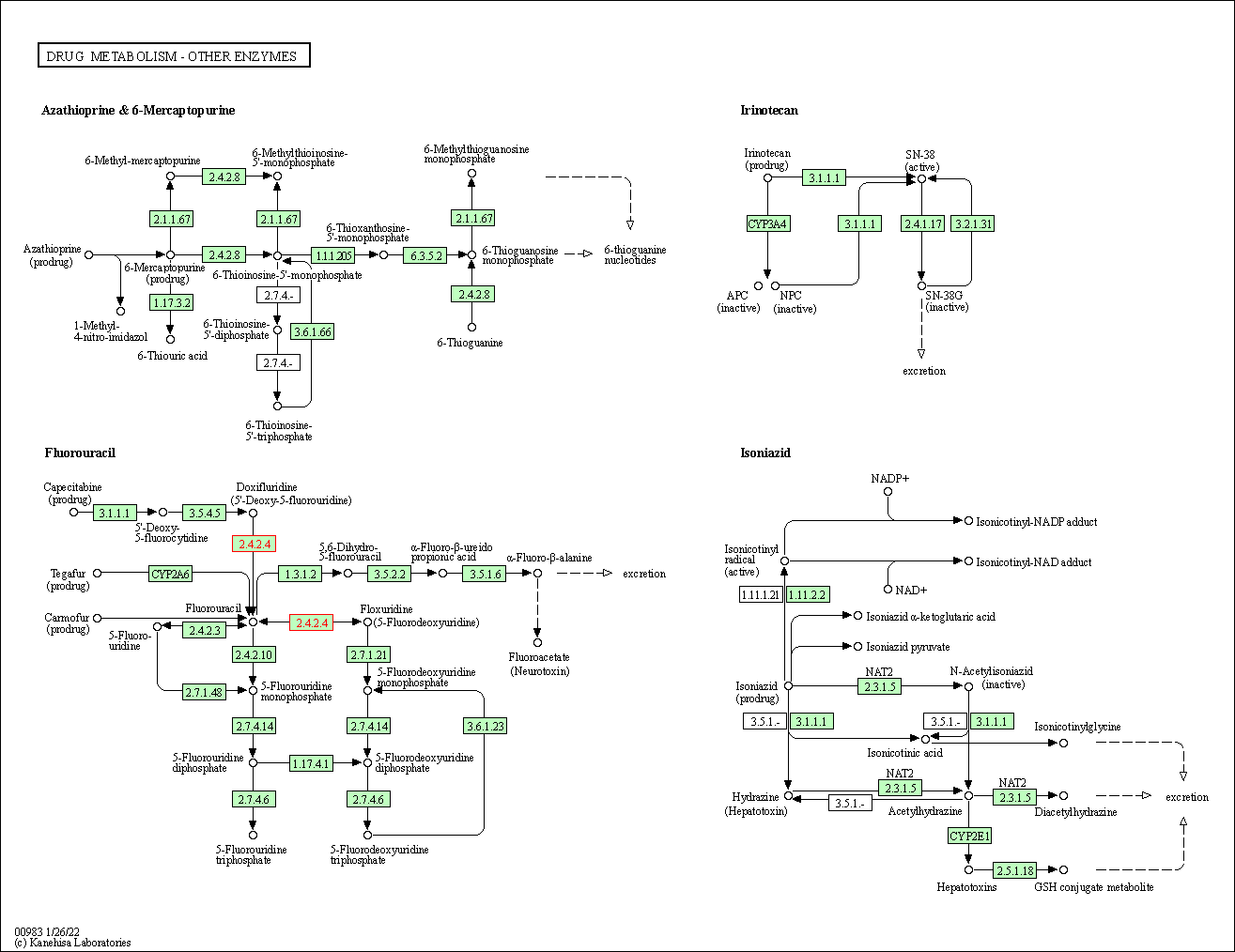
|
| Class: Metabolism => Xenobiotics biodegradation and metabolism | Pathway Hierarchy | ||
| Degree | 9 | Degree centrality | 9.67E-04 | Betweenness centrality | 6.05E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 1.70E-01 | Radiality | 1.27E+01 | Clustering coefficient | 3.33E-01 |
| Neighborhood connectivity | 7.56E+00 | Topological coefficient | 2.86E-01 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| BioCyc | [+] 1 BioCyc Pathways | + | ||||
| 1 | Pyrimidine deoxyribonucleosides degradation | |||||
| KEGG Pathway | [+] 4 KEGG Pathways | + | ||||
| 1 | Pyrimidine metabolism | |||||
| 2 | Drug metabolism - other enzymes | |||||
| 3 | Metabolic pathways | |||||
| 4 | Bladder cancer | |||||
| NetPath Pathway | [+] 1 NetPath Pathways | + | ||||
| 1 | TSH Signaling Pathway | |||||
| Pathwhiz Pathway | [+] 1 Pathwhiz Pathways | + | ||||
| 1 | Pyrimidine Metabolism | |||||
| WikiPathways | [+] 3 WikiPathways | + | ||||
| 1 | Bladder Cancer | |||||
| 2 | Metabolism of nucleotides | |||||
| 3 | Fluoropyrimidine Activity | |||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Enzymatic activities of uridine and thymidine phosphorylase in normal and cancerous uterine cervical tissues. Hum Cell. 2007 Nov;20(4):107-10. | |||||
| REF 2 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 4566). | |||||
| REF 3 | ClinicalTrials.gov (NCT00210080) A Study to Diagnose Lung Cancer by Sputum Cytology (01-312). U.S. National Institutes of Health. | |||||
| REF 4 | 5'-O-tritylinosine and analogues as allosteric inhibitors of human thymidine phosphorylase. J Med Chem. 2006 Sep 7;49(18):5562-70. | |||||
| REF 5 | Potential tumor-selective nitroimidazolylmethyluracil prodrug derivatives: inhibitors of the angiogenic enzyme thymidine phosphorylase. J Med Chem. 2003 Jan 16;46(2):207-9. | |||||
| REF 6 | Discovery of 5-substituted-6-chlorouracils as efficient inhibitors of human thymidine phosphorylase. J Med Chem. 2007 Nov 29;50(24):6016-23. | |||||
| REF 7 | Design and synthesis of novel 5,6-disubstituted uracil derivatives as potent inhibitors of thymidine phosphorylase. Bioorg Med Chem Lett. 2006 Mar 1;16(5):1335-7. | |||||
| REF 8 | The role of phosphate in the action of thymidine phosphorylase inhibitors: Implications for the catalytic mechanism. Bioorg Med Chem Lett. 2010 Mar 1;20(5):1648-51. | |||||
| REF 9 | Xanthine oxidase-activated prodrugs of thymidine phosphorylase inhibitors. Eur J Med Chem. 2008 Jun;43(6):1248-60. | |||||
| REF 10 | How many drug targets are there Nat Rev Drug Discov. 2006 Dec;5(12):993-6. | |||||
| REF 11 | Structural basis for non-competitive product inhibition in human thymidine phosphorylase: implications for drug design. Biochem J. 2006 Oct 15;399(2):199-204. | |||||
| REF 12 | Structures of native human thymidine phosphorylase and in complex with 5-iodouracil. Biochem Biophys Res Commun. 2009 Sep 4;386(4):666-70. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

