Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T11288
(Former ID: TTDS00288)
|
|||||
| Target Name |
Multidrug resistance-associated protein 1 (ABCC1)
|
|||||
| Synonyms |
MRP1; MRP; Leukotriene C(4) transporter; LTC4 transporter; ATP-binding cassette sub-family C member 1
Click to Show/Hide
|
|||||
| Gene Name |
ABCC1
|
|||||
| Target Type |
Successful target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Gout [ICD-11: FA25] | |||||
| Function |
Mediates ATP-dependent transport of glutathione and glutathione conjugates, leukotriene C4, estradiol-17-beta-o-glucuronide, methotrexate, antiviral drugs and other xenobiotics. Confers resistance to anticancer drugs. Hydrolyzes ATP with low efficiency. Mediates export of organic anions and drugs from the cytoplasm.
Click to Show/Hide
|
|||||
| BioChemical Class |
ABC transporter
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 7.6.2.2
|
|||||
| Sequence |
MALRGFCSADGSDPLWDWNVTWNTSNPDFTKCFQNTVLVWVPCFYLWACFPFYFLYLSRH
DRGYIQMTPLNKTKTALGFLLWIVCWADLFYSFWERSRGIFLAPVFLVSPTLLGITMLLA TFLIQLERRKGVQSSGIMLTFWLVALVCALAILRSKIMTALKEDAQVDLFRDITFYVYFS LLLIQLVLSCFSDRSPLFSETIHDPNPCPESSASFLSRITFWWITGLIVRGYRQPLEGSD LWSLNKEDTSEQVVPVLVKNWKKECAKTRKQPVKVVYSSKDPAQPKESSKVDANEEVEAL IVKSPQKEWNPSLFKVLYKTFGPYFLMSFFFKAIHDLMMFSGPQILKLLIKFVNDTKAPD WQGYFYTVLLFVTACLQTLVLHQYFHICFVSGMRIKTAVIGAVYRKALVITNSARKSSTV GEIVNLMSVDAQRFMDLATYINMIWSAPLQVILALYLLWLNLGPSVLAGVAVMVLMVPVN AVMAMKTKTYQVAHMKSKDNRIKLMNEILNGIKVLKLYAWELAFKDKVLAIRQEELKVLK KSAYLSAVGTFTWVCTPFLVALCTFAVYVTIDENNILDAQTAFVSLALFNILRFPLNILP MVISSIVQASVSLKRLRIFLSHEELEPDSIERRPVKDGGGTNSITVRNATFTWARSDPPT LNGITFSIPEGALVAVVGQVGCGKSSLLSALLAEMDKVEGHVAIKGSVAYVPQQAWIQND SLRENILFGCQLEEPYYRSVIQACALLPDLEILPSGDRTEIGEKGVNLSGGQKQRVSLAR AVYSNADIYLFDDPLSAVDAHVGKHIFENVIGPKGMLKNKTRILVTHSMSYLPQVDVIIV MSGGKISEMGSYQELLARDGAFAEFLRTYASTEQEQDAEENGVTGVSGPGKEAKQMENGM LVTDSAGKQLQRQLSSSSSYSGDISRHHNSTAELQKAEAKKEETWKLMEADKAQTGQVKL SVYWDYMKAIGLFISFLSIFLFMCNHVSALASNYWLSLWTDDPIVNGTQEHTKVRLSVYG ALGISQGIAVFGYSMAVSIGGILASRCLHVDLLHSILRSPMSFFERTPSGNLVNRFSKEL DTVDSMIPEVIKMFMGSLFNVIGACIVILLATPIAAIIIPPLGLIYFFVQRFYVASSRQL KRLESVSRSPVYSHFNETLLGVSVIRAFEEQERFIHQSDLKVDENQKAYYPSIVANRWLA VRLECVGNCIVLFAALFAVISRHSLSAGLVGLSVSYSLQVTTYLNWLVRMSSEMETNIVA VERLKEYSETEKEAPWQIQETAPPSSWPQVGRVEFRNYCLRYREDLDFVLRHINVTINGG EKVGIVGRTGAGKSSLTLGLFRINESAEGEIIIDGINIAKIGLHDLRFKITIIPQDPVLF SGSLRMNLDPFSQYSDEEVWTSLELAHLKDFVSALPDKLDHECAEGGENLSVGQRQLVCL ARALLRKTKILVLDEATAAVDLETDDLIQSTIRTQFEDCTVLTIAHRLNTIMDYTRVIVL DKGEIQEYGAPSDLLQQRGLFYSMAKDAGLV Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| HIT2.0 ID | T21EGX | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Approved Drug(s) | [+] 2 Approved Drugs | + | ||||
| 1 | Benzbromarone | Drug Info | Approved | Gout | [2] | |
| 2 | Sulfinpyrazone | Drug Info | Approved | Gout | [2], [3], [4] | |
| Mode of Action | [+] 3 Modes of Action | + | ||||
| Stimulator | [+] 1 Stimulator drugs | + | ||||
| 1 | Benzbromarone | Drug Info | [1] | |||
| Modulator | [+] 1 Modulator drugs | + | ||||
| 1 | Sulfinpyrazone | Drug Info | [5], [6], [7] | |||
| Inhibitor | [+] 5 Inhibitor drugs | + | ||||
| 1 | Pyrrolopyrimidine | Drug Info | [8] | |||
| 2 | XR-9456 | Drug Info | [9] | |||
| 3 | XR-9504 | Drug Info | [9] | |||
| 4 | XR-9544 | Drug Info | [9] | |||
| 5 | XR-9577 | Drug Info | [9] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Adenosine triphosphate | Ligand Info | |||||
| Structure Description | Structure of the human Multidrug Resistance Protein 1 Nucleotide Binding Domain 1 | PDB:2CBZ | ||||
| Method | X-ray diffraction | Resolution | 1.50 Å | Mutation | No | [10] |
| PDB Sequence |
NSITVRNATF
651 TWARSDPPTL661 NGITFSIPEG671 ALVAVVGQVG681 CGKSSLLSAL691 LAEMDKVEGH 701 VAIKGSVAYV711 PQQAWIQNDS721 LRENILFGCQ731 LEEPYYRSVI741 QACALLPDLE 751 ILPSGDRTEI761 GEKGVNLSGG771 QKQRVSLARA781 VYSNADIYLF791 DDPLSAVDAH 801 VGKHIFENVI811 GPKGMLKNKT821 RILVTHSMSY831 LPQVDVIIVM841 SGGKISEMGS 851 YQELLARDGA861 FAEFLRTYAS871 H
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| ABC transporters | hsa02010 | Affiliated Target |
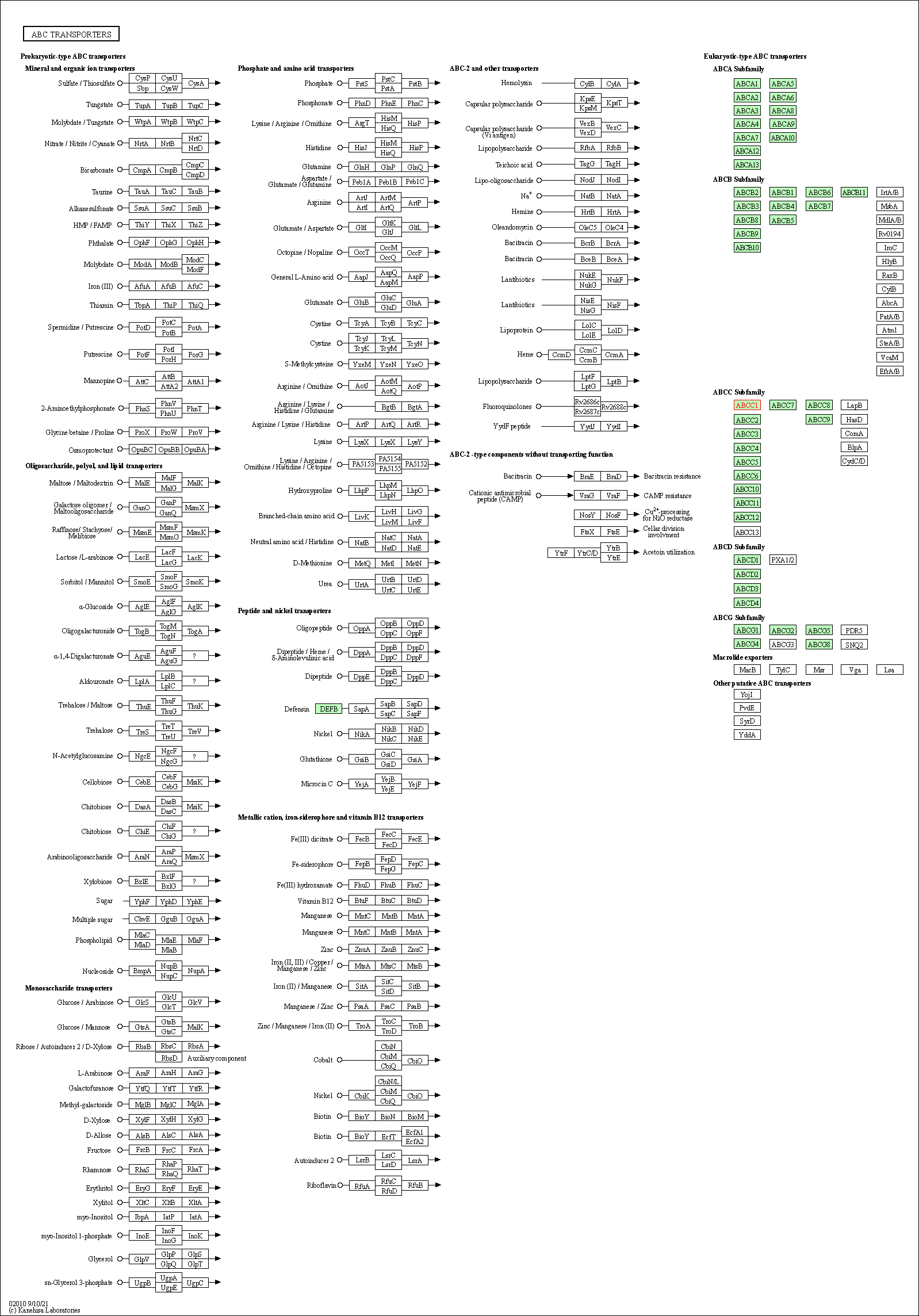
|
| Class: Environmental Information Processing => Membrane transport | Pathway Hierarchy | ||
| Sphingolipid signaling pathway | hsa04071 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Vitamin digestion and absorption | hsa04977 | Affiliated Target |
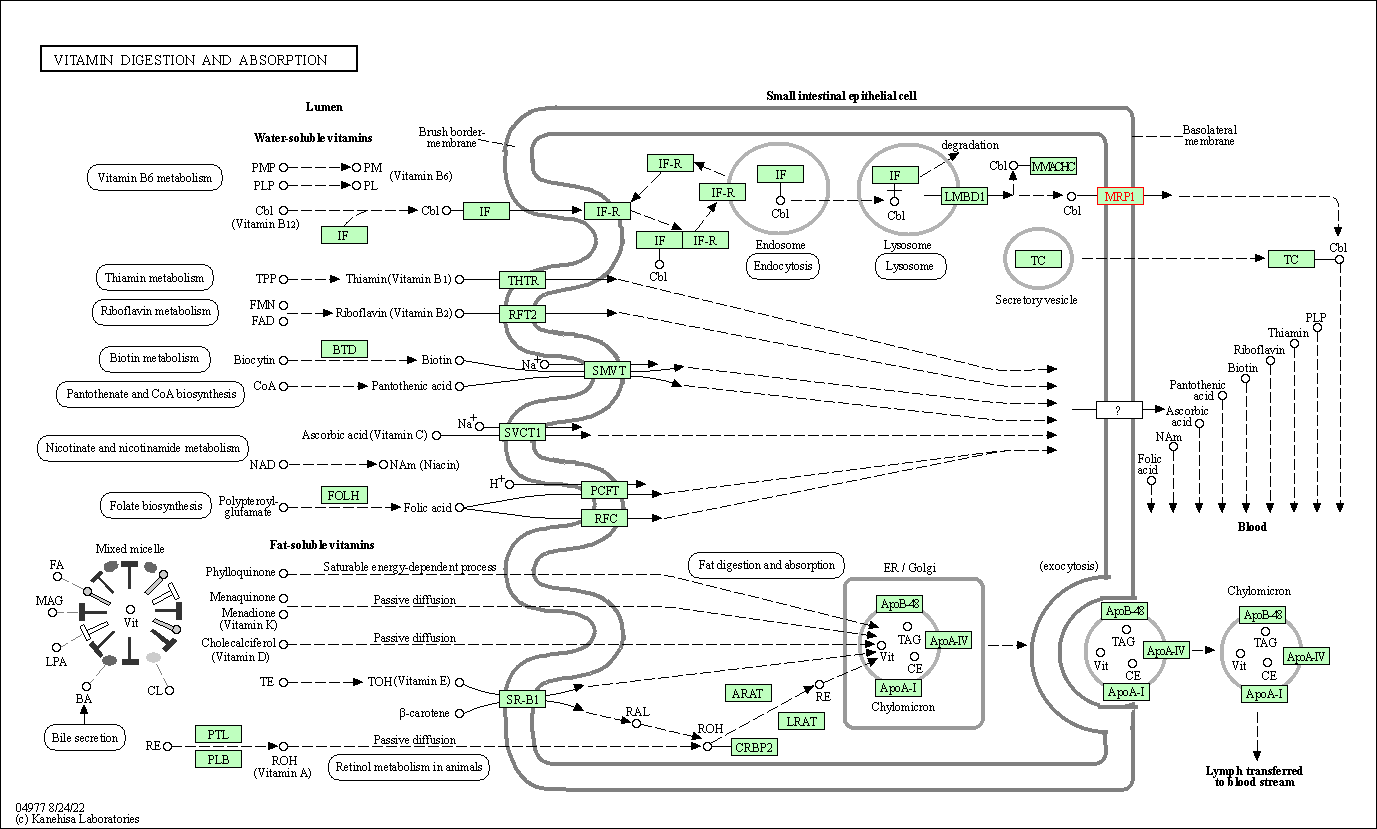
|
| Class: Organismal Systems => Digestive system | Pathway Hierarchy | ||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 4 KEGG Pathways | + | ||||
| 1 | ABC transporters | |||||
| 2 | Sphingolipid signaling pathway | |||||
| 3 | Vitamin digestion and absorption | |||||
| 4 | MicroRNAs in cancer | |||||
| NetPath Pathway | [+] 1 NetPath Pathways | + | ||||
| 1 | IL6 Signaling Pathway | |||||
| PID Pathway | [+] 2 PID Pathways | + | ||||
| 1 | S1P1 pathway | |||||
| 2 | Sphingosine 1-phosphate (S1P) pathway | |||||
| Reactome | [+] 2 Reactome Pathways | + | ||||
| 1 | Cobalamin (Cbl, vitamin B12) transport and metabolism | |||||
| 2 | ABC-family proteins mediated transport | |||||
| WikiPathways | [+] 3 WikiPathways | + | ||||
| 1 | Arachidonic acid metabolism | |||||
| 2 | Irinotecan Pathway | |||||
| 3 | Metabolism of water-soluble vitamins and cofactors | |||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Transport of diclofenac by breast cancer resistance protein (ABCG2) and stimulation of multidrug resistance protein 2 (ABCC2)-mediated drug transpo... Drug Metab Dispos. 2009 Jan;37(1):129-36. | |||||
| REF 2 | Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services. 2015 | |||||
| REF 3 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 5826). | |||||
| REF 4 | FDA Approved Drug Products from FDA Official Website. 2009. Application Number: (NDA) 011556. | |||||
| REF 5 | Species-dependent transport and modulation properties of human and mouse multidrug resistance protein 2 (MRP2/Mrp2, ABCC2/Abcc2). Drug Metab Dispos. 2008 Apr;36(4):631-40. | |||||
| REF 6 | Mutation of a single conserved tryptophan in multidrug resistance protein 1 (MRP1/ABCC1) results in loss of drug resistance and selective loss of o... J Biol Chem. 2001 May 11;276(19):15616-24. | |||||
| REF 7 | Genome duplications and other features in 12 Mb of DNA sequence from human chromosome 16p and 16q. Genomics. 1999 Sep 15;60(3):295-308. | |||||
| REF 8 | Topological model for the prediction of MRP1 inhibitory activity of pyrrolopyrimidines and templates derived from pyrrolopyrimidine. Bioorg Med Chem Lett. 2005 Nov 15;15(22):4967-72. | |||||
| REF 9 | Functional assay and structure-activity relationships of new third-generation P-glycoprotein inhibitors. Bioorg Med Chem. 2008 Mar 1;16(5):2448-62. | |||||
| REF 10 | Structure of the human multidrug resistance protein 1 nucleotide binding domain 1 bound to Mg2+/ATP reveals a non-productive catalytic site. J Mol Biol. 2006 Jun 16;359(4):940-9. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

