Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T63024
(Former ID: TTDI03570)
|
|||||
| Target Name |
NF-kappa-B-activating kinase (TBK1)
|
|||||
| Synonyms |
TANK-binding kinase 1; T2K; Serine/threonine-protein kinase TBK1; NAK
Click to Show/Hide
|
|||||
| Gene Name |
TBK1
|
|||||
| Target Type |
Patented-recorded target
|
[1] | ||||
| Function |
Following activation of toll-like receptors by viral or bacterial components, associates with TRAF3 and TANK and phosphorylates interferon regulatory factors (IRFs) IRF3 and IRF7 as well as DDX3X. This activity allows subsequent homodimerization and nuclear translocation of the IRFs leading to transcriptional activation of pro-inflammatory and antiviral genes including IFNA and IFNB. In order to establish such an antiviral state, TBK1 form several different complexes whose composition depends on the type of cell and cellular stimuli. Plays a key role in IRF3 activation: acts by first phosphorylating innate adapter proteins MAVS, TMEM173/STING and TICAM1 on their pLxIS motif, leading to recruitment of IRF3, thereby licensing IRF3 for phosphorylation by TBK1. Phosphorylated IRF3 dissociates from the adapter proteins, dimerizes, and then enters the nucleus to induce expression of interferons. Thus, several scaffolding molecules including FADD, TRADD, MAVS, AZI2, TANK or TBKBP1/SINTBAD can be recruited to the TBK1-containing-complexes. Under particular conditions, functions as a NF-kappa-B effector by phosphorylating NF-kappa-B inhibitor alpha/NFKBIA, IKBKB or RELA to translocate NF-Kappa-B to the nucleus. Restricts bacterial proliferation by phosphorylating the autophagy receptor OPTN/Optineurin on 'Ser-177', thus enhancing LC3 binding affinity and antibacterial autophagy. Phosphorylates SMCR8 component of the C9orf72-SMCR8 complex, promoting autophagosome maturation. Phosphorylates and activates AKT1. Seems to play a role in energy balance regulation by sustaining a state of chronic, low-grade inflammation in obesity, wich leads to a negative impact on insulin sensitivity. Attenuates retroviral budding by phosphorylating the endosomal sorting complex required for transport-I (ESCRT-I) subunit VPS37C. Phosphorylates Borna disease virus (BDV) P protein. Plays an essential role in the TLR3- and IFN-dependent control of herpes virus HSV-1 and HSV-2 infections in the central nervous system. Serine/threonine kinase that plays an essential role in regulating inflammatory responses to foreign agents.
Click to Show/Hide
|
|||||
| BioChemical Class |
Kinase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.7.11.1
|
|||||
| Sequence |
MQSTSNHLWLLSDILGQGATANVFRGRHKKTGDLFAIKVFNNISFLRPVDVQMREFEVLK
KLNHKNIVKLFAIEEETTTRHKVLIMEFCPCGSLYTVLEEPSNAYGLPESEFLIVLRDVV GGMNHLRENGIVHRDIKPGNIMRVIGEDGQSVYKLTDFGAARELEDDEQFVSLYGTEEYL HPDMYERAVLRKDHQKKYGATVDLWSIGVTFYHAATGSLPFRPFEGPRRNKEVMYKIITG KPSGAISGVQKAENGPIDWSGDMPVSCSLSRGLQVLLTPVLANILEADQEKCWGFDQFFA ETSDILHRMVIHVFSLQQMTAHKIYIHSYNTATIFHELVYKQTKIISSNQELIYEGRRLV LEPGRLAQHFPKTTEENPIFVVSREPLNTIGLIYEKISLPKVHPRYDLDGDASMAKAITG VVCYACRIASTLLLYQELMRKGIRWLIELIKDDYNETVHKKTEVVITLDFCIRNIEKTVK VYEKLMKINLEAAELGEISDIHTKLLRLSSSQGTIETSLQDIDSRLSPGGSLADAWAHQE GTHPKDRNVEKLQVLLNCMTEIYYQFKKDKAERRLAYNEEQIHKFDKQKLYYHATKAMTH FTDECVKKYEAFLNKSEEWIRKMLHLRKQLLSLTNQCFDIEEEVSKYQEYTNELQETLPQ KMFTASSGIKHTMTPIYPSSNTLVEMTLGMKKLKEEMEGVVKELAENNHILERFGSLTMD GGLRNVDCL Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| HIT2.0 ID | T60GGR | |||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Amlexanox | Ligand Info | |||||
| Structure Description | TBK1 co-crystal structure with amlexanox | PDB:5W5V | ||||
| Method | X-ray diffraction | Resolution | 3.65 Å | Mutation | No | [4] |
| PDB Sequence |
MQSTSNHLWL
10 LSDILTANVF24 RGRHKKTGDL34 FAIKVFVDVQ52 MREFEVLKKL62 NHKNIVKLFA 72 IEEETTTRHK82 VLIMEFCPCG92 SLYTVLEEPS102 NAYGLPESEF112 LIVLRDVVGG 122 MNHLRENGIV132 HRDIKPGNIM142 RVIGEDGQSV152 YKLTDFGEEY179 LHPDMYERAT 201 VDLWSIGVTF211 YHAATGSLPF221 RPFEGPRRNK231 EVMYKIITGK241 PSGAISGVQK 251 AENGPIDWSG261 DMPVSCSLSR271 GLQVLLTPVL281 ANILDQEKCW293 GFDQFFAETS 303 DILHRMVIHV313 FSLQQMTAHK323 IYIHSYNTAT333 IFHELVYKQT343 KIISSNQELI 353 YEGRRLVLEP363 GRLAQHFPKT373 TEENPIFVVS383 REPLNTIGLI393 YEKISLPKVH 403 PRYDLDGDAS413 MAKAITGVVC423 YACRIASTLL433 LYQELMRKGI443 RWLIELIKDD 453 YNETVHKKTE463 VVITLDFCIR473 NIEKTVKVEI498 SDIHTKLLRL508 SSSQGTIETS 518 LQDIDSRLSP528 GGSLADAWAH538 QEGTHPKDRN548 VEKLQVLLNC558 MTEIYYQFKK 568 DKAERRLAYN578 EEQIHKFDKQ588 KLYYHATKAM598 THFTDECVKK608 YEAFLNKSEE 618 WIRKMLHLRK628 QLLSLTNQCF638 DIEEEVSKYQ648 EYTNELQET
|
|||||
|
|
||||||
| Ligand Name: MRT67307 | Ligand Info | |||||
| Structure Description | Structure of Tank-Binding Kinase 1 | PDB:4IM0 | ||||
| Method | X-ray diffraction | Resolution | 2.40 Å | Mutation | Yes | [5] |
| PDB Sequence |
GSMQSTSNHL
8 WLLSDILGQG18 ATANVFRGRH28 KKTGDLFAIK38 VFNNRPVDVQ52 MREFEVLKKL 62 NHKNIVKLFA72 IEEETTTRHK82 VLIMEFCPCG92 SLYTVLEEPS102 NAYGLPESEF 112 LIVLRDVVGG122 MNHLRENGIV132 HRNIKPGNIM142 RVIGEDGQSV152 YKLTDFGTEE 178 YLHPDMYERV189 LRKDHATVDL204 WSIGVTFYHA214 ATGSLPFRPF224 EGPRRNKEVM 234 YKIITGKPSG244 AISGVQKAEN254 GPIDWSGDMP264 VSCSLSRGLQ274 VLLTPVLANI 284 LEADQEKCWG294 FDQFFAETSD304 ILHRMVIHVF314 SLQQMTAHKI324 YIHSYNTATI 334 FHELVYKQTK344 IISSNQELIY354 EGRRLVLEPG364 RLAQHFPKTT374 EENPIFVVSR 384 EPLNTIGLIY394 EKISLPKVHP404 RYDLDGDASM414 AKAITGVVCY424 ACRIASTLLL 434 YQELMRKGIR444 WLIELIKDDY454 NETVHKKTEV464 VITLDFCIRN474 IEKTVKVMEI 498 SDIHTKLLRL508 SSSQGTIETS518 LQDIDSRLSP528 GGSLADAWAH538 QEGTHPKDRN 548 VEKLQVLLNC558 MTEIYYQFKK568 DKAERRLAYN578 EEQIHKFDKQ588 KLYYHATKAM 598 THFTDECVKK608 YEAFLNKSEE618 WIRKMLHLRK628 QLLSLTNQCF638 DIEEEVSKYQ 648 EYTNELQET
|
|||||
|
|
LEU15
3.675
GLY16
4.180
GLN17
3.930
GLY18
3.655
ALA21
4.163
VAL23
3.474
ARG25
4.501
ALA36
3.560
LYS38
3.715
VAL68
3.606
MET86
3.591
|
|||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Ras signaling pathway | hsa04014 | Affiliated Target |
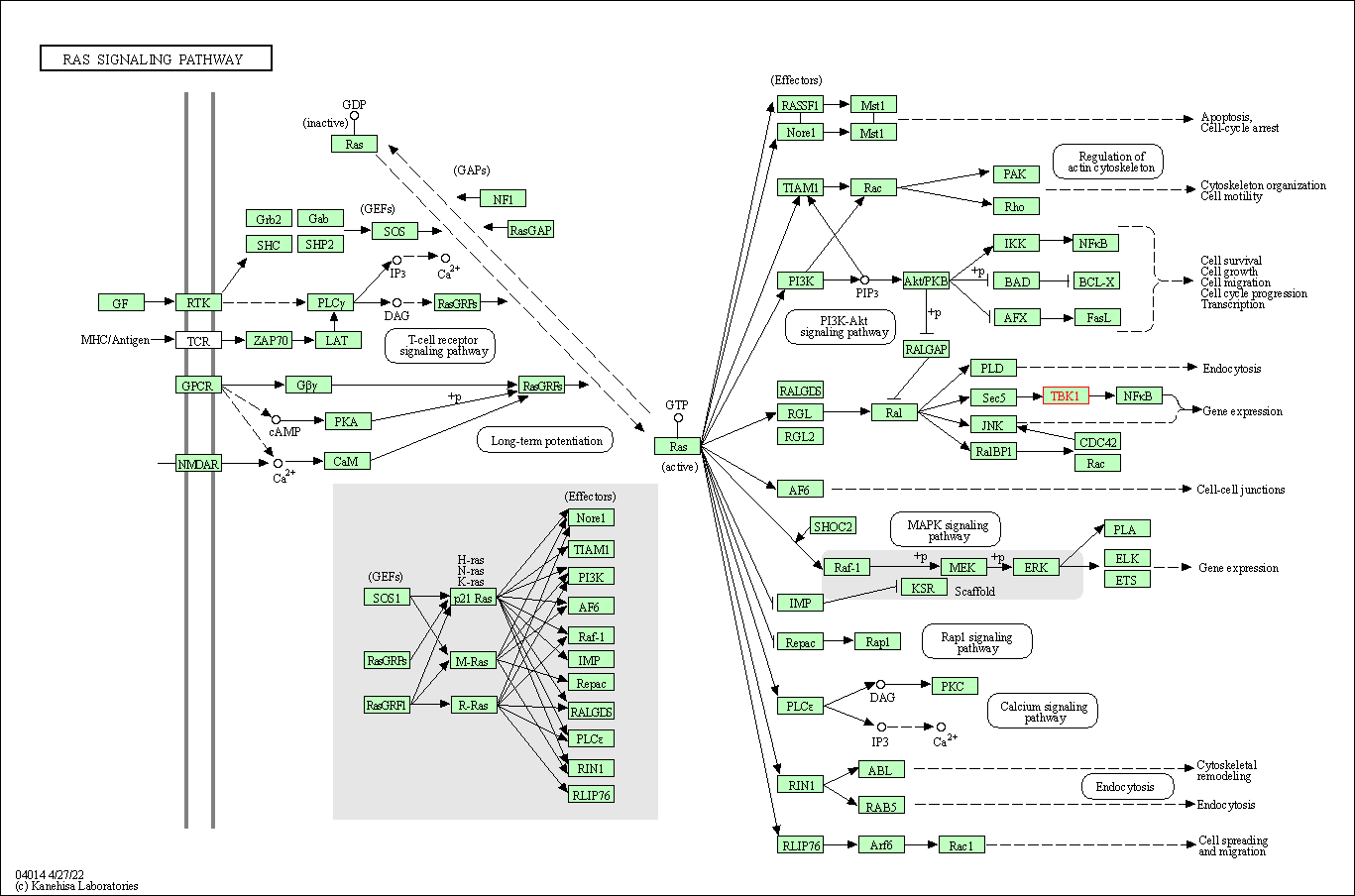
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Mitophagy - animal | hsa04137 | Affiliated Target |
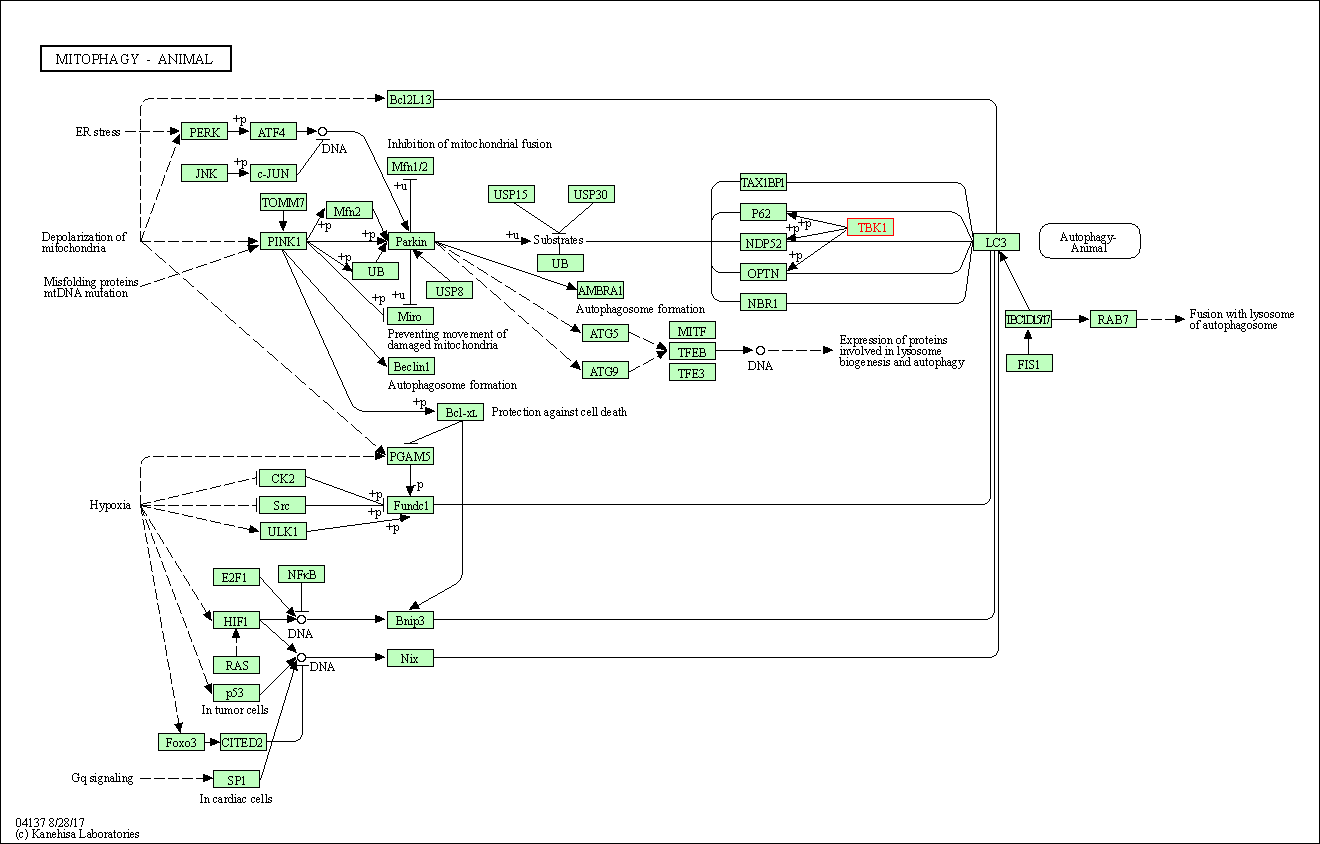
|
| Class: Cellular Processes => Transport and catabolism | Pathway Hierarchy | ||
| Autophagy - animal | hsa04140 | Affiliated Target |
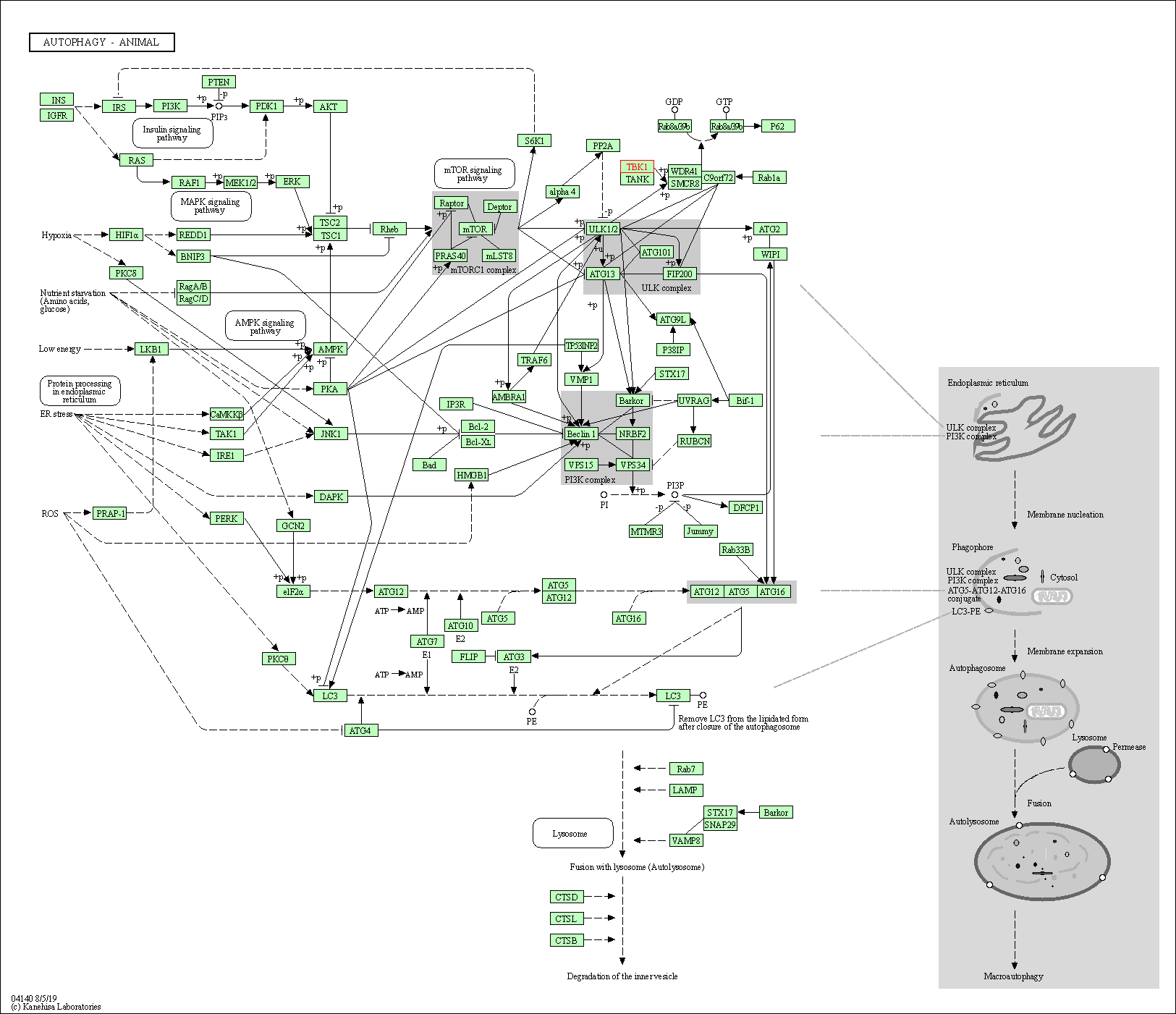
|
| Class: Cellular Processes => Transport and catabolism | Pathway Hierarchy | ||
| Toll-like receptor signaling pathway | hsa04620 | Affiliated Target |
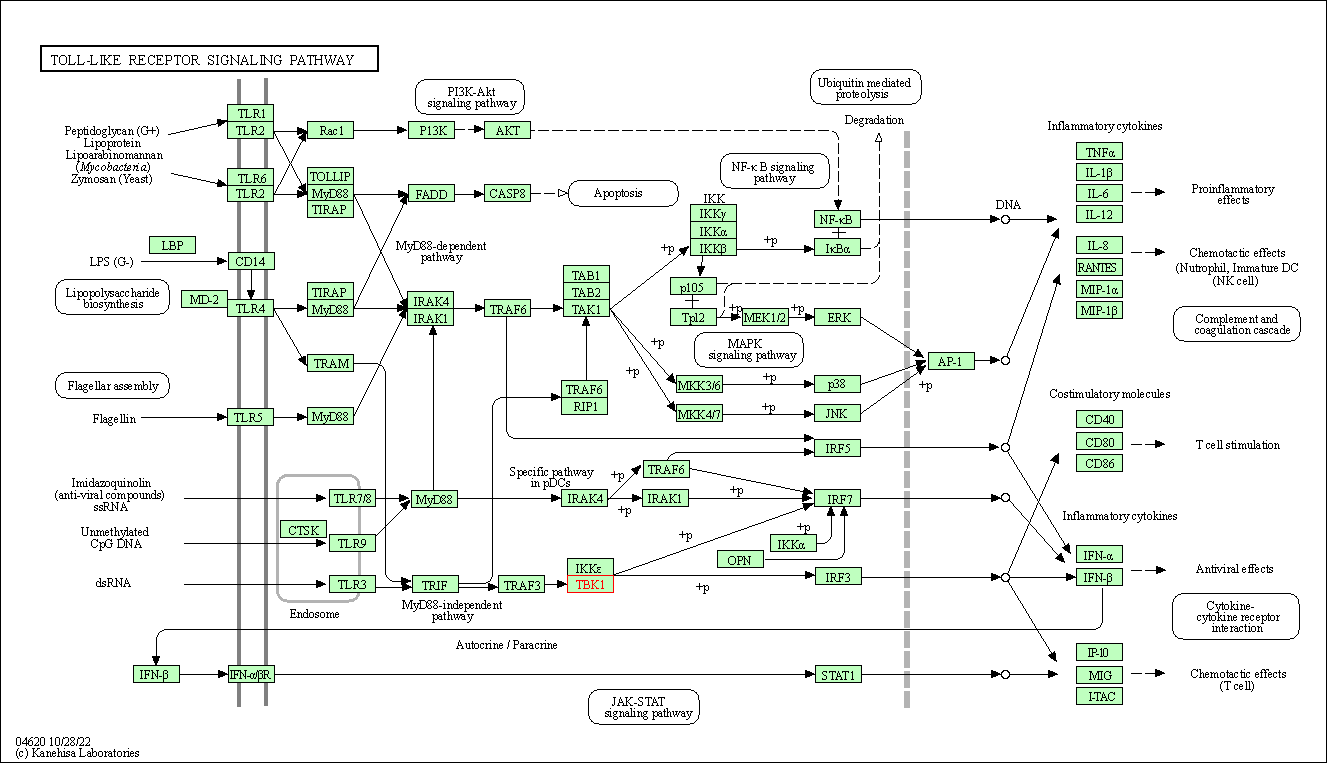
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| NOD-like receptor signaling pathway | hsa04621 | Affiliated Target |
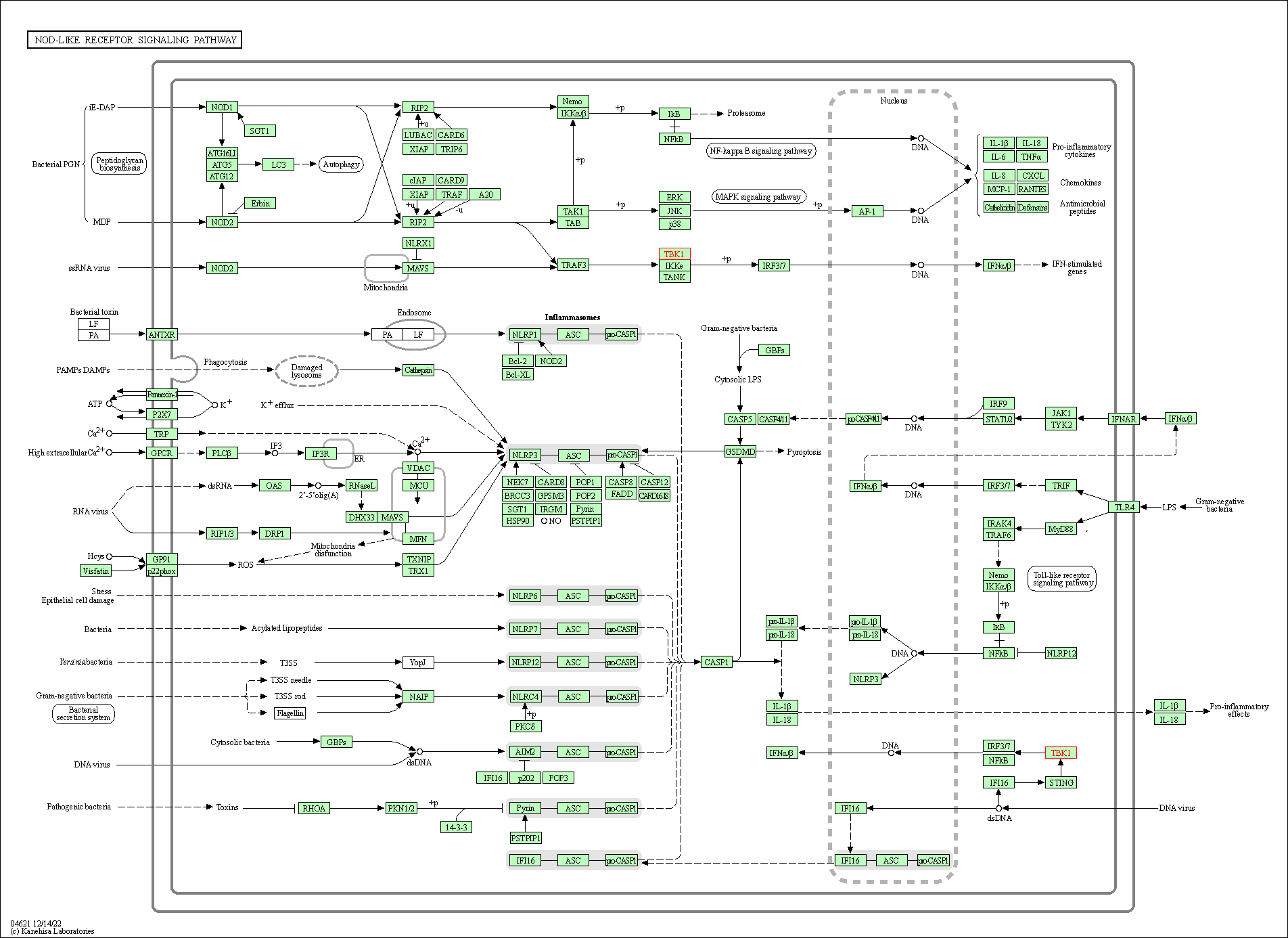
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| RIG-I-like receptor signaling pathway | hsa04622 | Affiliated Target |
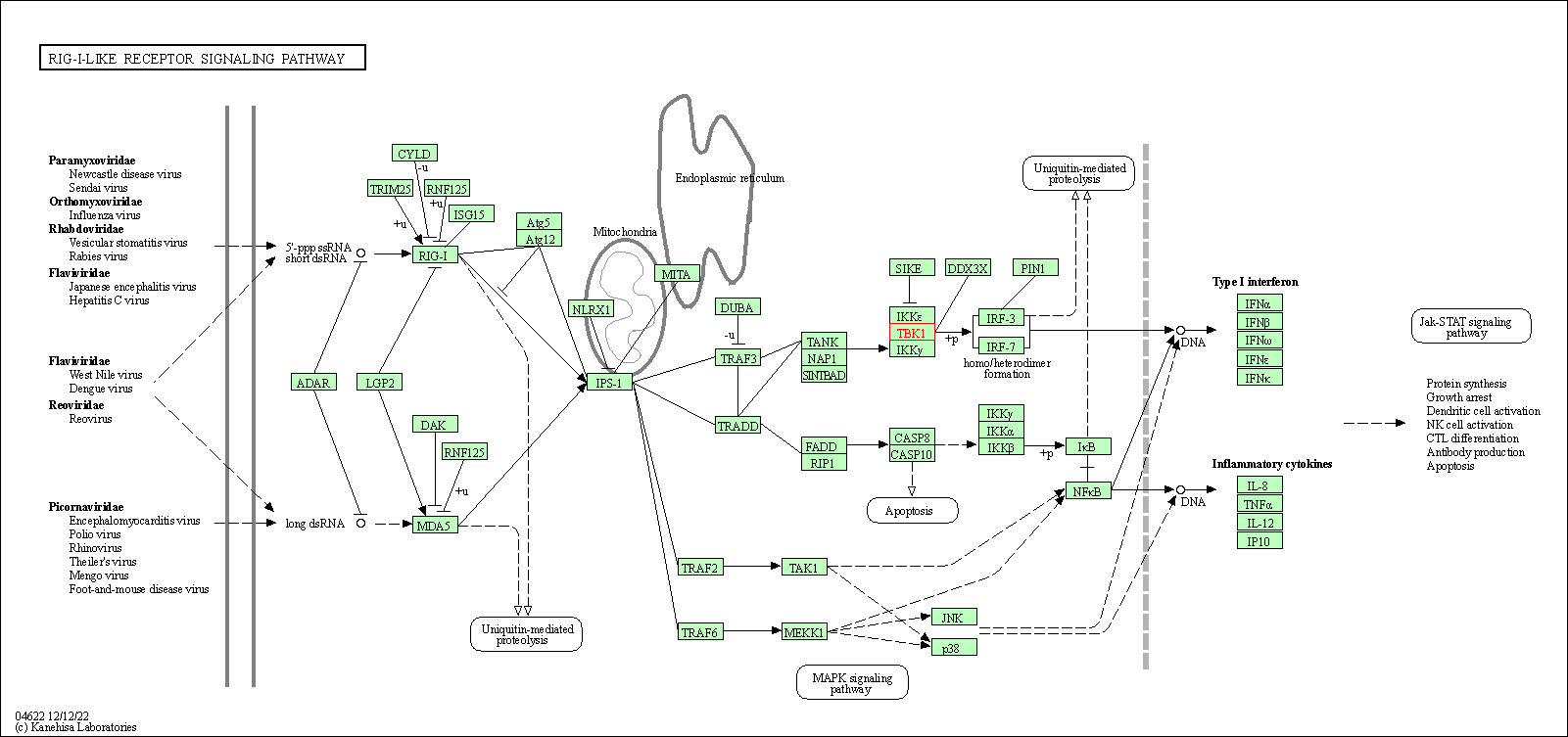
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Cytosolic DNA-sensing pathway | hsa04623 | Affiliated Target |
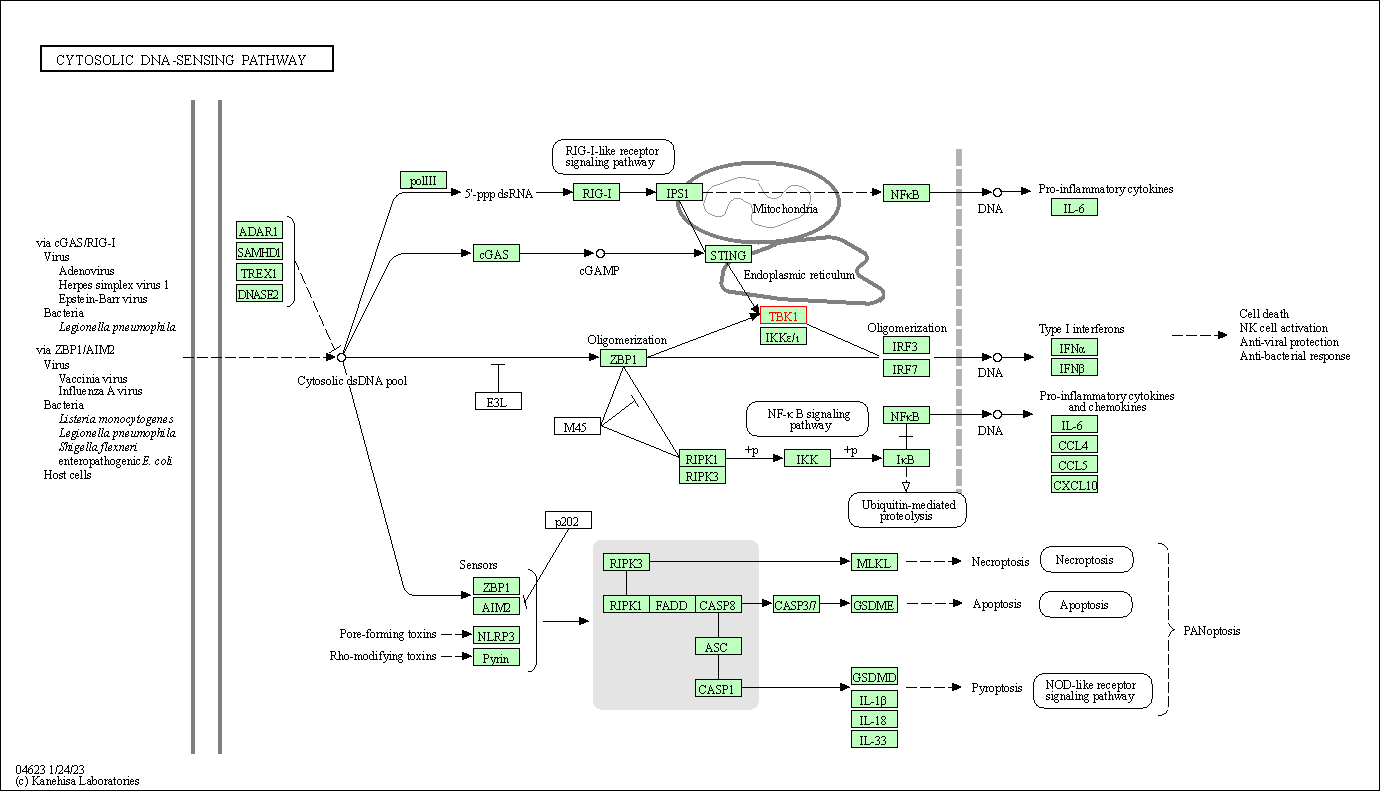
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| IL-17 signaling pathway | hsa04657 | Affiliated Target |
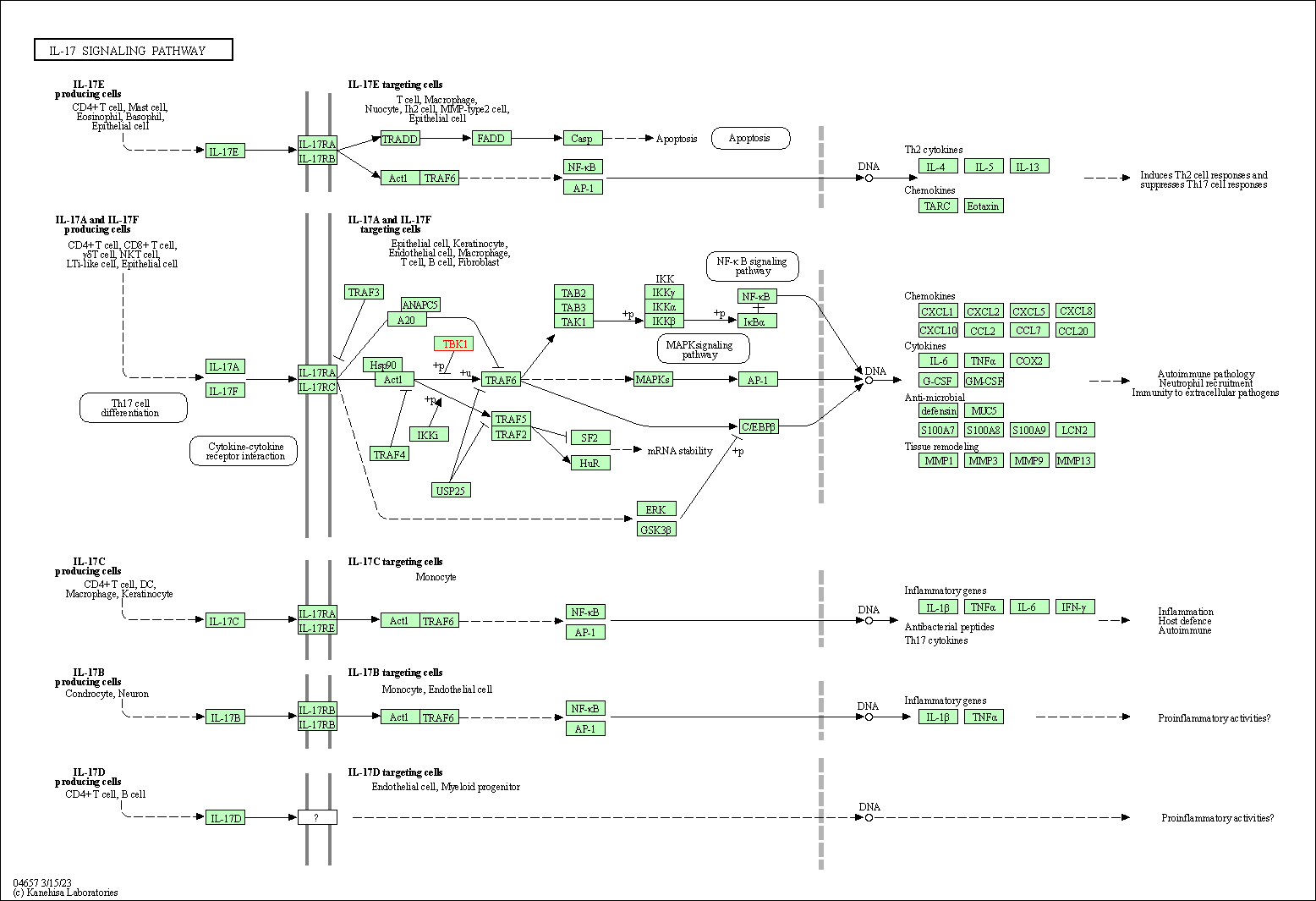
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 29 | Degree centrality | 3.12E-03 | Betweenness centrality | 1.27E-03 |
|---|---|---|---|---|---|
| Closeness centrality | 2.42E-01 | Radiality | 1.42E+01 | Clustering coefficient | 1.80E-01 |
| Neighborhood connectivity | 3.36E+01 | Topological coefficient | 6.07E-02 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | TBK1 inhibitors: a review of patent literature (2011 - 2014).Expert Opin Ther Pat. 2015;25(12):1385-96. | |||||
| REF 2 | Novel cross-talk within the IKK family controls innate immunity. Biochem J. 2011 Feb 15;434(1):93-104. | |||||
| REF 3 | Synthesis and structure-activity relationships of a novel series of pyrimidines as potent inhibitors of TBK1/IKKepsilon kinases. Bioorg Med Chem Lett. 2012 Dec 1;22(23):7169-73. | |||||
| REF 4 | Carboxylic Acid Derivatives of Amlexanox Display Enhanced Potency toward TBK1 and IKKEpsilon and Reveal Mechanisms for Selective Inhibition. Mol Pharmacol. 2018 Oct;94(4):1210-1219. | |||||
| REF 5 | Structure and ubiquitination-dependent activation of TANK-binding kinase 1. Cell Rep. 2013 Mar 28;3(3):747-58. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

