Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T62841
(Former ID: TTDR00109)
|
|||||
| Target Name |
Proteinase activated receptor 2 (PAR2)
|
|||||
| Synonyms |
Thrombin receptor-like 1; Proteinase-activated receptor-2; Proteinase-activated receptor 2; Protease activated receptor 2; PAR-2; GPR11; G-protein coupled receptor 11; Coagulation factor II receptor-like 1
Click to Show/Hide
|
|||||
| Gene Name |
F2RL1
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Chronic pain [ICD-11: MG30] | |||||
| Function |
Its function is mediated through the activation of several signaling pathways including phospholipase C (PLC), intracellular calcium, mitogen-activated protein kinase (MAPK), I-kappaB kinase/NF-kappaB and Rho. Can also be transactivated by cleaved F2R/PAR1. Involved in modulation of inflammatory responses and regulation of innate and adaptive immunity, and acts as a sensor for proteolytic enzymes generated during infection. Generally is promoting inflammation. Can signal synergistically with TLR4 and probably TLR2 in inflammatory responses and modulates TLR3 signaling. Has a protective role in establishing the endothelial barrier; the activity involves coagulation factor X. Regulates endothelial cell barrier integrity during neutrophil extravasation, probably following proteolytic cleavage by PRTN3. Proposed to have a bronchoprotective role in airway epithelium, but also shown to compromise the airway epithelial barrier by interrupting E-cadherin adhesion. Involved in the regulation of vascular tone; activation results in hypotension presumably mediated by vasodilation. Associates with a subset of G proteins alpha subunits such as GNAQ, GNA11, GNA14, GNA12 and GNA13, but probably not with G(o) alpha, G(i) subunit alpha-1 and G(i) subunit alpha-2. However, according to can signal through G(i) subunit alpha. Believed to be a class B receptor which internalizes as a complex with arrestin and traffic with it to endosomal vesicles, presumably as desensitized receptor, for extended periods of time. Mediates inhibition of TNF-alpha stimulated JNK phosphorylation via coupling to GNAQ and GNA11; the function involves dissociation of RIPK1 and TRADD from TNFR1. Mediates phosphorylation of nuclear factor NF-kappa-B RELA subunit at 'Ser-536'; the function involves IKBKB and is predominantly independent of G proteins. Involved in cellular migration. Involved in cytoskeletal rearrangement and chemotaxis through beta-arrestin-promoted scaffolds; the function is independent of GNAQ and GNA11 and involves promotion of cofilin dephosphorylation and actin filament severing. Induces redistribution of COPS5 from the plasma membrane to the cytosol and activation of the JNK cascade is mediated by COPS5. Involved in the recruitment of leukocytes to the sites of inflammation and is the major PAR receptor capable of modulating eosinophil function such as proinflammatory cytokine secretion, superoxide production and degranulation. During inflammation promotes dendritic cell maturation, trafficking to the lymph nodes and subsequent T-cell activation. Involved in antimicrobial response of innate immnune cells; activation enhances phagocytosis of Gram-positive and killing of Gram-negative bacteria. Acts synergistically with interferon-gamma in enhancing antiviral responses. Implicated in a number of acute and chronic inflammatory diseases such as of the joints, lungs, brain, gastrointestinal tract, periodontium, skin, and vascular systems, and in autoimmune disorders. Receptor for trypsin and trypsin-like enzymes coupled to G proteins.
Click to Show/Hide
|
|||||
| BioChemical Class |
GPCR rhodopsin
|
|||||
| UniProt ID | ||||||
| Sequence |
MRSPSAAWLLGAAILLAASLSCSGTIQGTNRSSKGRSLIGKVDGTSHVTGKGVTVETVFS
VDEFSASVLTGKLTTVFLPIVYTIVFVVGLPSNGMALWVFLFRTKKKHPAVIYMANLALA DLLSVIWFPLKIAYHIHGNNWIYGEALCNVLIGFFYGNMYCSILFMTCLSVQRYWVIVNP MGHSRKKANIAIGISLAIWLLILLVTIPLYVVKQTIFIPALNITTCHDVLPEQLLVGDMF NYFLSLAIGVFLFPAFLTASAYVLMIRMLRSSAMDENSEKKRKRAIKLIVTVLAMYLICF TPSNLLLVVHYFLIKSQGQSHVYALYIVALCLSTLNSCIDPFVYYFVSHDFRDHAKNALL CRSVRTVKQMQVSLTSKKHSRKSSSYSSSSTTVKTSY Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| HIT2.0 ID | T55WN8 | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 1 Clinical Trial Drugs | + | ||||
| 1 | MEDI0618 | Drug Info | Phase 1 | Osteoarthritis pain | [2] | |
| Patented Agent(s) | [+] 29 Patented Agents | + | ||||
| 1 | Amidine compound 1 | Drug Info | Patented | Inflammation | [1] | |
| 2 | Amidine compound 2 | Drug Info | Patented | Inflammation | [1] | |
| 3 | Amidine compound 3 | Drug Info | Patented | Inflammation | [1] | |
| 4 | Amidine compound 4 | Drug Info | Patented | Inflammation | [1] | |
| 5 | Amidine compound 5 | Drug Info | Patented | Inflammation | [1] | |
| 6 | Amidine compound 6 | Drug Info | Patented | Inflammation | [1] | |
| 7 | Benzothiazine-carboxamide compound 1 | Drug Info | Patented | Skin inflammation | [1] | |
| 8 | Benzothiazine-carboxamide compound 2 | Drug Info | Patented | Skin inflammation | [1] | |
| 9 | Benzothiazine-carboxamide compound 3 | Drug Info | Patented | Skin inflammation | [1] | |
| 10 | Benzothiazine-carboxamide compound 4 | Drug Info | Patented | Skin inflammation | [1] | |
| 11 | Benzothiazine-carboxamide compound 5 | Drug Info | Patented | Skin inflammation | [1] | |
| 12 | Benzothiazine-carboxamide compound 6 | Drug Info | Patented | Skin inflammation | [1] | |
| 13 | Imidazopyridazine derivative 3 | Drug Info | Patented | Pain | [1] | |
| 14 | Imidazopyridazine derivative 4 | Drug Info | Patented | Pain | [1] | |
| 15 | Imidazopyridazine derivative 5 | Drug Info | Patented | Pain | [1] | |
| 16 | Imidazopyridazine derivative 6 | Drug Info | Patented | Pain | [1] | |
| 17 | Imidazopyridazine derivative 7 | Drug Info | Patented | Pain | [1] | |
| 18 | Imidazopyridazine derivative 8 | Drug Info | Patented | Pain | [1] | |
| 19 | Piperazine derivative 5 | Drug Info | Patented | Atopic dermatitis | [1] | |
| 20 | Piperazine urea derivative 3 | Drug Info | Patented | Inflammation | [1] | |
| 21 | Piperazine urea derivative 4 | Drug Info | Patented | Inflammation | [1] | |
| 22 | Piperidine derivative 2 | Drug Info | Patented | Inflammation | [1] | |
| 23 | Piperidine derivative 3 | Drug Info | Patented | Inflammation | [1] | |
| 24 | PMID26936077-Compound-13 | Drug Info | Patented | Type-2 diabetes | [1] | |
| 25 | PMID26936077-Compound-34 | Drug Info | Patented | Type-2 diabetes | [1] | |
| 26 | Quinazoline derivative 10 | Drug Info | Patented | Cirrhosis | [1] | |
| 27 | Quinazoline derivative 11 | Drug Info | Patented | Cirrhosis | [1] | |
| 28 | Quinazoline derivative 12 | Drug Info | Patented | Cirrhosis | [1] | |
| 29 | Quinazoline derivative 13 | Drug Info | Patented | Multiple sclerosis | [1] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Antagonist | [+] 34 Antagonist drugs | + | ||||
| 1 | MEDI0618 | Drug Info | [3] | |||
| 2 | Amidine compound 1 | Drug Info | [1] | |||
| 3 | Amidine compound 2 | Drug Info | [1] | |||
| 4 | Amidine compound 3 | Drug Info | [1] | |||
| 5 | Amidine compound 4 | Drug Info | [1] | |||
| 6 | Amidine compound 5 | Drug Info | [1] | |||
| 7 | Amidine compound 6 | Drug Info | [1] | |||
| 8 | Benzimidazole derivative 10 | Drug Info | [1] | |||
| 9 | Benzimidazole derivative 8 | Drug Info | [1] | |||
| 10 | Benzimidazole derivative 9 | Drug Info | [1] | |||
| 11 | Benzothiazine-carboxamide compound 1 | Drug Info | [1] | |||
| 12 | Benzothiazine-carboxamide compound 2 | Drug Info | [1] | |||
| 13 | Benzothiazine-carboxamide compound 3 | Drug Info | [1] | |||
| 14 | Benzothiazine-carboxamide compound 4 | Drug Info | [1] | |||
| 15 | Benzothiazine-carboxamide compound 5 | Drug Info | [1] | |||
| 16 | Benzothiazine-carboxamide compound 6 | Drug Info | [1] | |||
| 17 | Imidazopyridazine derivative 3 | Drug Info | [1] | |||
| 18 | Imidazopyridazine derivative 4 | Drug Info | [1] | |||
| 19 | Imidazopyridazine derivative 5 | Drug Info | [1] | |||
| 20 | Imidazopyridazine derivative 6 | Drug Info | [1] | |||
| 21 | Imidazopyridazine derivative 7 | Drug Info | [1] | |||
| 22 | Imidazopyridazine derivative 8 | Drug Info | [1] | |||
| 23 | Piperazine derivative 5 | Drug Info | [1] | |||
| 24 | Piperazine urea derivative 3 | Drug Info | [1] | |||
| 25 | Piperazine urea derivative 4 | Drug Info | [1] | |||
| 26 | Piperidine derivative 2 | Drug Info | [1] | |||
| 27 | Piperidine derivative 3 | Drug Info | [1] | |||
| 28 | PMID26936077-Compound-13 | Drug Info | [1] | |||
| 29 | PMID26936077-Compound-34 | Drug Info | [1] | |||
| 30 | Quinazoline derivative 10 | Drug Info | [1] | |||
| 31 | Quinazoline derivative 11 | Drug Info | [1] | |||
| 32 | Quinazoline derivative 12 | Drug Info | [1] | |||
| 33 | Quinazoline derivative 13 | Drug Info | [1] | |||
| 34 | GB88 | Drug Info | [6] | |||
| Agonist | [+] 5 Agonist drugs | + | ||||
| 1 | Peptide analog 71 | Drug Info | [1] | |||
| 2 | Peptide analog 72 | Drug Info | [1] | |||
| 3 | Peptidomimetic analog 6 | Drug Info | [1] | |||
| 4 | ASKH95 | Drug Info | [4] | |||
| 5 | GB110 | Drug Info | [5] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
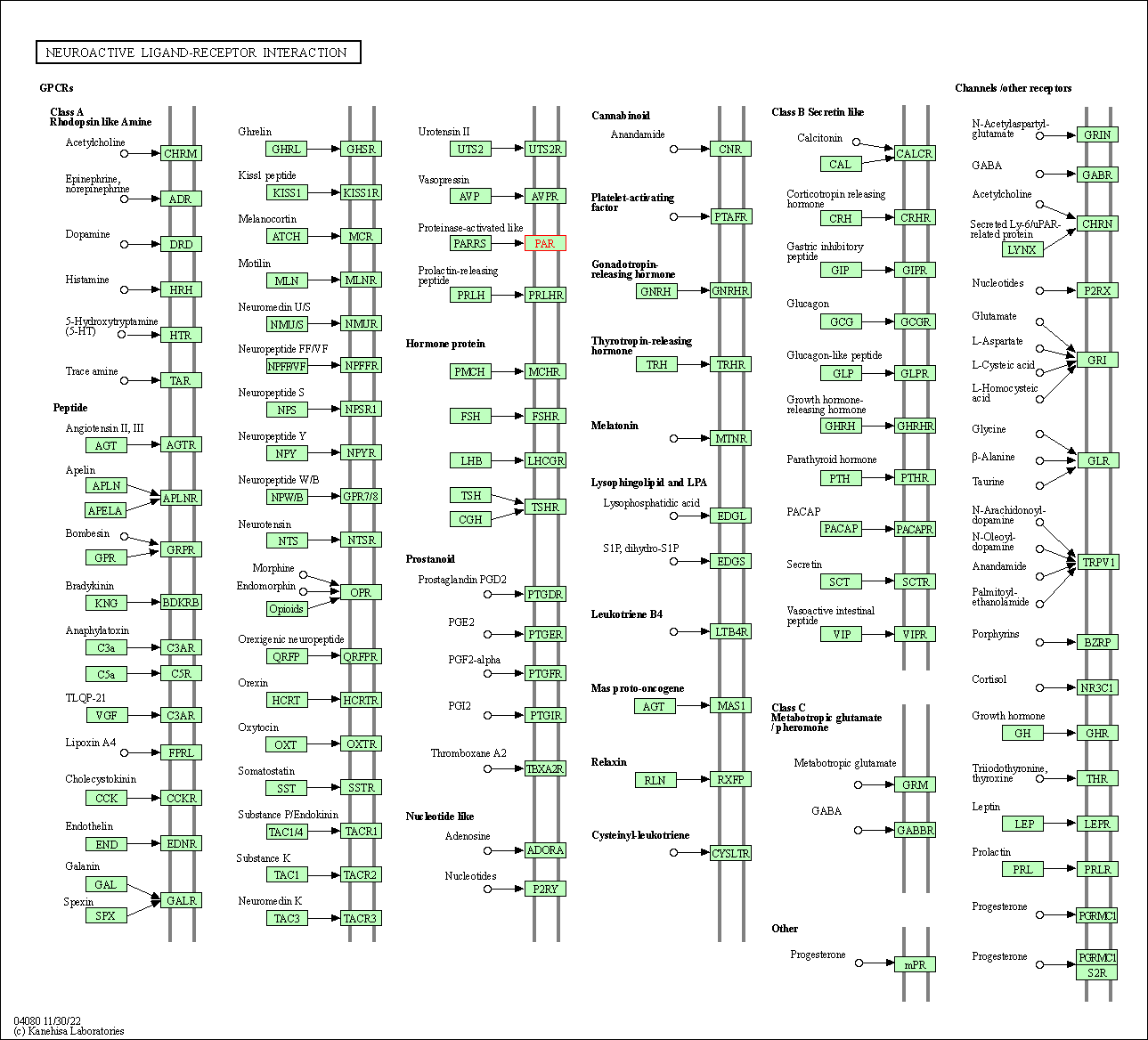
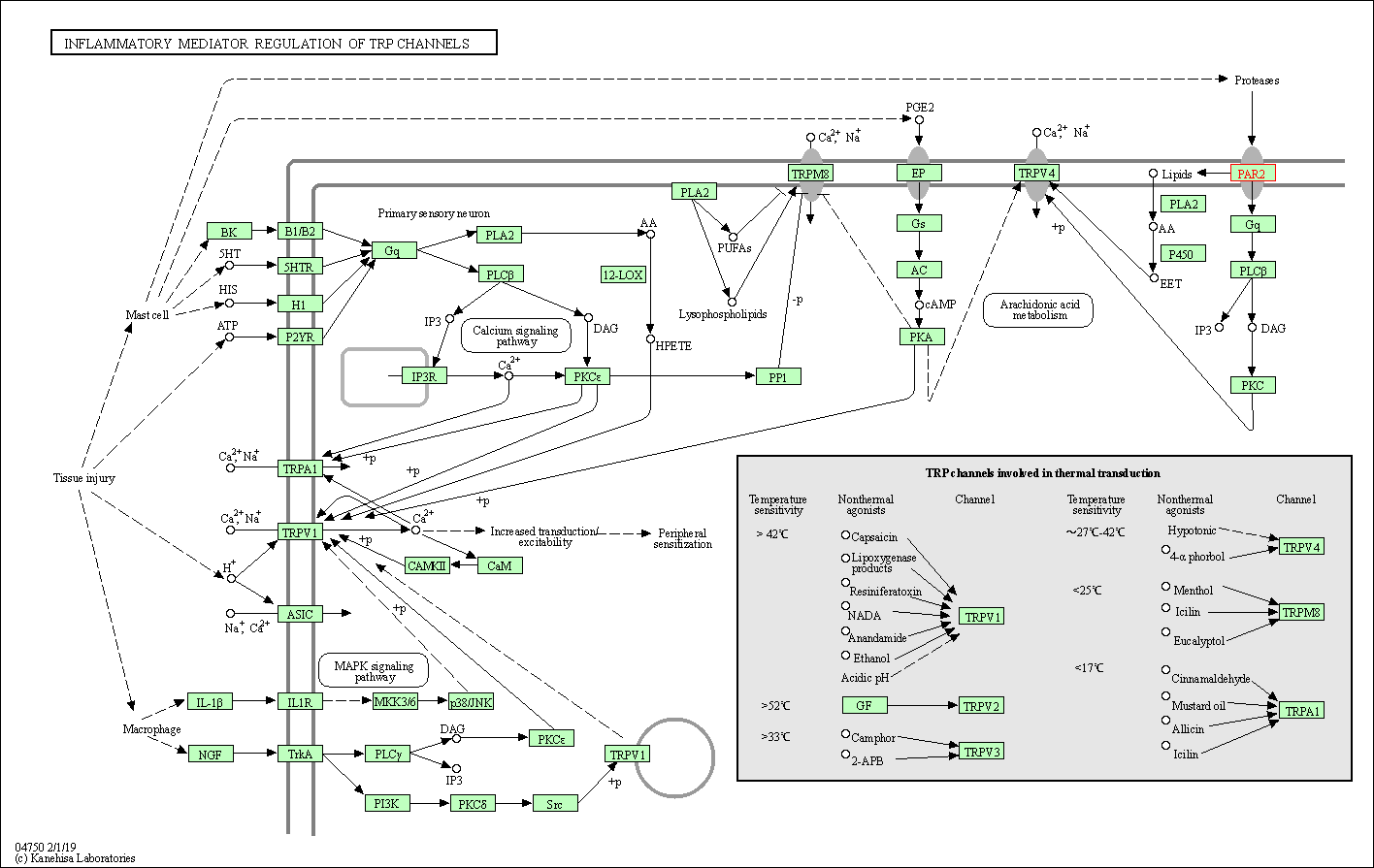
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Neuroactive ligand-receptor interaction | hsa04080 | Affiliated Target |

|
| Class: Environmental Information Processing => Signaling molecules and interaction | Pathway Hierarchy | ||
| Inflammatory mediator regulation of TRP channels | hsa04750 | Affiliated Target |

|
| Class: Organismal Systems => Sensory system | Pathway Hierarchy | ||
| Degree | 1 | Degree centrality | 1.07E-04 | Betweenness centrality | 0.00E+00 |
|---|---|---|---|---|---|
| Closeness centrality | 1.86E-01 | Radiality | 1.31E+01 | Clustering coefficient | 0.00E+00 |
| Neighborhood connectivity | 5.20E+01 | Topological coefficient | 1.00E+00 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 3 KEGG Pathways | + | ||||
| 1 | Neuroactive ligand-receptor interaction | |||||
| 2 | Inflammatory mediator regulation of TRP channels | |||||
| 3 | African trypanosomiasis | |||||
| Reactome | [+] 2 Reactome Pathways | + | ||||
| 1 | Peptide ligand-binding receptors | |||||
| 2 | G alpha (q) signalling events | |||||
| WikiPathways | [+] 4 WikiPathways | + | ||||
| 1 | GPCRs, Class A Rhodopsin-like | |||||
| 2 | Gastrin-CREB signalling pathway via PKC and MAPK | |||||
| 3 | GPCR ligand binding | |||||
| 4 | GPCR downstream signaling | |||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Protease activated receptor 2 (PAR2) modulators: a patent review (2010-2015).Expert Opin Ther Pat. 2016;26(4):471-83. | |||||
| REF 2 | ClinicalTrials.gov (NCT05714254) A Randomised, Double-blind, Placebo-controlled Study of the Safety, Tolerability, and Pharmacokinetics of Multiple Ascending Doses of MEDI0618 in Healthy Male and Female Volunteers. U.S.National Institutes of Health. | |||||
| REF 3 | Clinical pipeline report, company report or official report of AstraZeneca | |||||
| REF 4 | Essential role for proteinase-activated receptor-2 in arthritis. J Clin Invest. 2003 Jan;111(1):35-41. | |||||
| REF 5 | Novel agonists and antagonists for human protease activated receptor 2. J Med Chem. 2010 Oct 28;53(20):7428-40. | |||||
| REF 6 | Modulating human proteinase activated receptor 2 with a novel antagonist (GB88) and agonist (GB110). Br J Pharmacol. 2012 Mar;165(5):1413-23. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

