Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T39321
(Former ID: TTDR00995)
|
|||||
| Target Name |
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH)
|
|||||
| Synonyms |
Peptidyl-cysteine S-nitrosylase GAPDH; OK/SW-cl.12; GAPD; D-glyceraldehyde-3-phosphate dehydrogenase; CDABP0047
Click to Show/Hide
|
|||||
| Gene Name |
GAPDH
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 2 Target-related Diseases | + | ||||
| 1 | Muscular atrophy [ICD-11: 8B61] | |||||
| 2 | Muscular dystrophy [ICD-11: 8C70] | |||||
| Function |
Participates in nuclear events including transcription, RNA transport, DNA replication and apoptosis. Nuclear functions are probably due to the nitrosylase activity that mediates cysteine S-nitrosylation of nuclear target proteins such as SIRT1, HDAC2 and PRKDC. Modulates the organization and assembly of the cytoskeleton. Facilitates the CHP1-dependent microtubule and membrane associations through its ability to stimulate the binding of CHP1 to microtubules. Glyceraldehyde-3-phosphate dehydrogenase is a key enzyme in glycolysis that catalyzes the first step of the pathway by converting D-glyceraldehyde 3-phosphate (G3P) into 3-phospho-D-glyceroyl phosphate. Component of the GAIT (gamma interferon-activated inhibitor of translation) complex which mediates interferon-gamma-induced transcript-selective translation inhibition in inflammation processes. Upon interferon-gamma treatment assembles into the GAIT complex which binds to stem loop-containing GAIT elements in the 3'-UTR of diverse inflammatory mRNAs (such as ceruplasmin) and suppresses their translation. Has both glyceraldehyde-3-phosphate dehydrogenase and nitrosylase activities, thereby playing a role in glycolysis and nuclear functions, respectively.
Click to Show/Hide
|
|||||
| BioChemical Class |
Aldehyde/oxo donor oxidoreductase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 1.2.1.12
|
|||||
| Sequence |
MGKVKVGVNGFGRIGRLVTRAAFNSGKVDIVAINDPFIDLNYMVYMFQYDSTHGKFHGTV
KAENGKLVINGNPITIFQERDPSKIKWGDAGAEYVVESTGVFTTMEKAGAHLQGGAKRVI ISAPSADAPMFVMGVNHEKYDNSLKIISNASCTTNCLAPLAKVIHDNFGIVEGLMTTVHA ITATQKTVDGPSGKLWRDGRGALQNIIPASTGAAKAVGKVIPELNGKLTGMAFRVPTANV SVVDLTCRLEKPAKYDDIKKVVKQASEGPLKGILGYTEHQVVSSDFNSDTHSSTFDAGAG IALNDHFVKLISWYDNEFGYSNRVVDLMAHMASKE Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| ADReCS ID | BADD_A06414 | |||||
| HIT2.0 ID | T23Z2G | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 1 Clinical Trial Drugs | + | ||||
| 1 | Omigapil | Drug Info | Phase 2 | Lateral sclerosis | [2] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Modulator | [+] 1 Modulator drugs | + | ||||
| 1 | Omigapil | Drug Info | [1] | |||
| Binder | [+] 1 Binder drugs | + | ||||
| 1 | Saframycin A | Drug Info | [3] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: S-sulpho-L-cysteine | Ligand Info | |||||
| Structure Description | GAPDH purified from the supernatant of HEK293F cells: crystal form 1 of 4. | PDB:6YND | ||||
| Method | X-ray diffraction | Resolution | 1.52 Å | Mutation | No | [4] |
| PDB Sequence |
GKVKVGVNGF
11 GRIGRLVTRA21 AFNSGKVDIV31 AINDPFIDLN41 YMVYMFQYDS51 THGKFHGTVK 61 AENGKLVING71 NPITIFQERD81 PSKIKWGDAG91 AEYVVESTGV101 FTTMEKAGAH 111 LQGGAKRVII121 SAPSADAPMF131 VMGVNHEKYD141 NSLKIISNAS151 TTNCLAPLAK 162 VIHDNFGIVE172 GLMTTVHAIT182 ATQKTVDGPS192 GKLWRDGRGA202 LQNIIPASTG 212 AAKAVGKVIP222 ELNGKLTGMA232 FRVPTANVSV242 VDLTCRLEKP252 AKYDDIKKVV 262 KQASEGPLKG272 ILGYTEHQVV282 SSDFNSDTHS292 STFDAGAGIA302 LNDHFVKLIS 312 WYDNEFGYSN322 RVVDLMAHMA332 SKE
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Ligand Name: L-serine-O-phosphate | Ligand Info | |||||
| Structure Description | Crystal structure of phosphorylated mutant of glyceraldehyde 3-phosphate dehydrogenase from human placenta at 3.15A resolution | PDB:6ADE | ||||
| Method | X-ray diffraction | Resolution | 3.15 Å | Mutation | Yes | [5] |
| PDB Sequence |
KVKVGVNGFG
12 RIGRLVTRAA22 FNSGKVDIVA32 INDPFIDLNY42 MVYMFQYDTH53 GKFHGTVKAE 63 NGKLVINGNP73 ITIFQERDPS83 KIKWGDAGAE93 YVVESTGVFT103 TMEKAGAHLQ 113 GGAKRVIISA123 PSADAPMFVM133 GVNHEKYDNS143 LKIISNACTT154 NCLAPLAKVI 164 HDNFGIVEGL174 MTTVHAITAT184 QKTVDGPSGK194 LWRDGRGALQ204 NIIPASTGAA 214 KAVGKVIPEL224 NGKLTGMAFR234 VPTANVSVVD244 LTCRLEKPAK254 YDDIKKVVKQ 264 ASEGPLLGIL274 GYTEHQVVSS284 DFNSDTHSST294 FDAGAGIALN304 DHFVKLISWY 314 DNEFGYSNRV324 VDLMAHMASK334 E
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Glycolysis / Gluconeogenesis | hsa00010 | Affiliated Target |
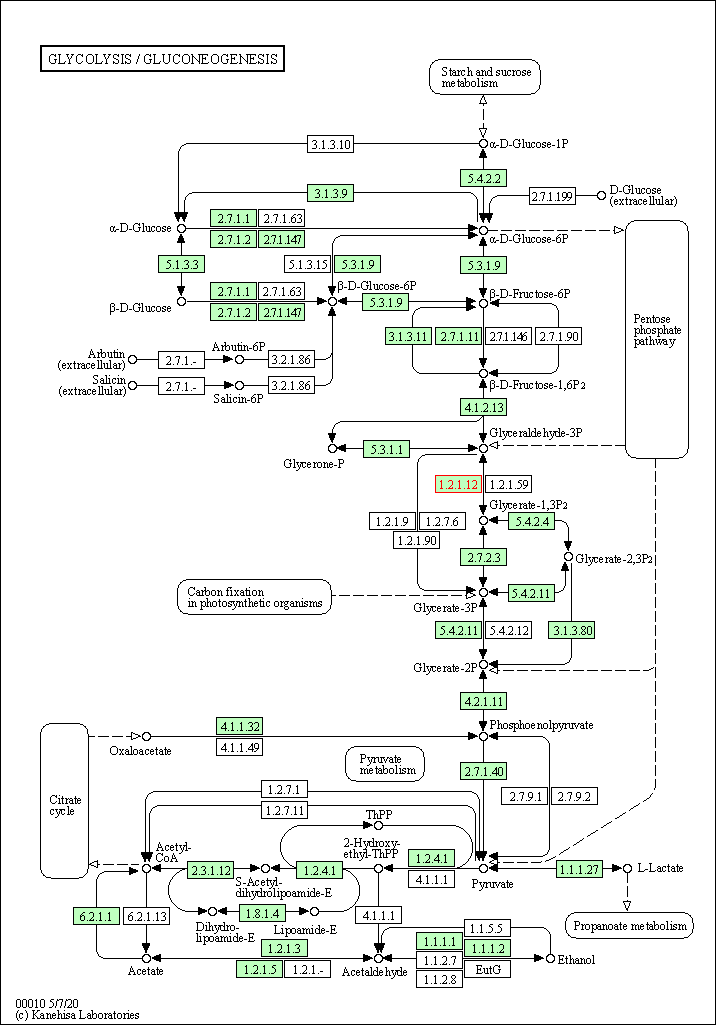
|
| Class: Metabolism => Carbohydrate metabolism | Pathway Hierarchy | ||
| HIF-1 signaling pathway | hsa04066 | Affiliated Target |
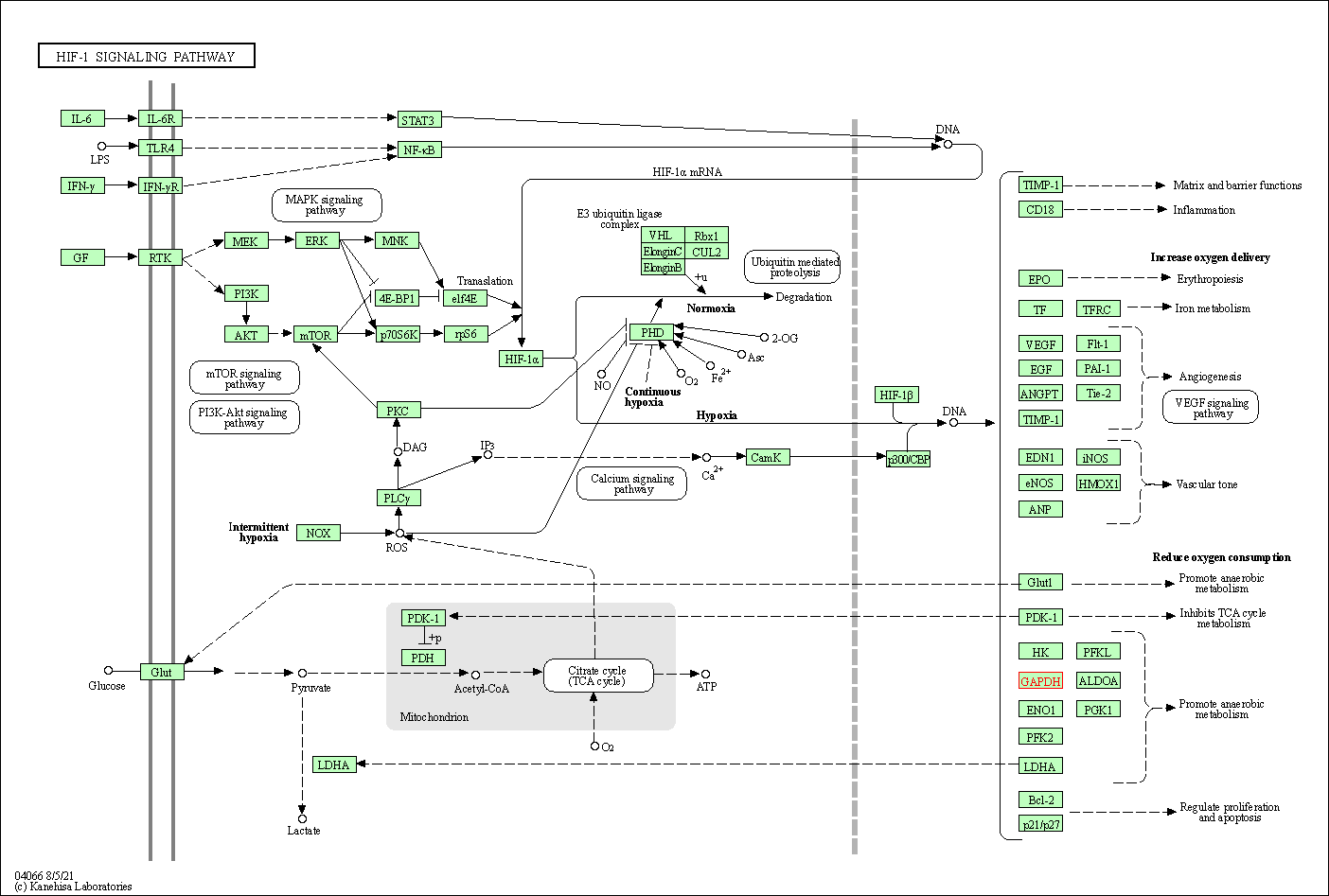
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Degree | 32 | Degree centrality | 3.44E-03 | Betweenness centrality | 1.19E-02 |
|---|---|---|---|---|---|
| Closeness centrality | 2.56E-01 | Radiality | 1.44E+01 | Clustering coefficient | 9.68E-02 |
| Neighborhood connectivity | 3.03E+01 | Topological coefficient | 4.18E-02 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Neuroprotection by pharmacologic blockade of the GAPDH death cascade. Proc Natl Acad Sci U S A. 2006 Mar 7;103(10):3887-9. | |||||
| REF 2 | ClinicalTrials.gov (NCT00649961) Melatonin As A Novel Neuroprotectant In Preterm Infants- Dosage Study. U.S. National Institutes of Health. | |||||
| REF 3 | Identification of GAPDH as a protein target of the saframycin antiproliferative agents. Proc Natl Acad Sci U S A. 2004 Apr 20;101(16):5862-6. | |||||
| REF 4 | Partial catalytic Cys oxidation of human GAPDH to Cys-sulfonic acid. Wellcome Open Res. 2020 Aug 25;5:114. | |||||
| REF 5 | Crystal structure of phosphorylated mutant of glyceraldehyde 3-phosphate dehydrogenase from human placenta at 3.15A resolution | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

