Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T93798
(Former ID: TTDI03120)
|
|||||
| Target Name |
Cyclic nucleotide-gated channel alpha-1 (CNGA1)
|
|||||
| Synonyms |
cGMP-gated cation channel alpha-1; Rod photoreceptor cGMP-gated channel subunit alpha; Cyclic nucleotide-gated channel, photoreceptor; Cyclic nucleotide-gated cation channel 1; CNG1; CNG-1; CNG channel alpha-1; CNCG1; CNCG
Click to Show/Hide
|
|||||
| Gene Name |
CNGA1
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Myelopathy [ICD-11: 8B42] | |||||
| Function |
Visual signal transduction is mediated by a G-protein coupled cascade using cGMP as second messenger. This protein can be activated by cyclic GMP which leads to an opening of the cation channel and thereby causing a depolarization of rod photoreceptors.
Click to Show/Hide
|
|||||
| UniProt ID | ||||||
| Sequence |
MKLSMKNNIINTQQSFVTMPNVIVPDIEKEIRRMENGACSSFSEDDDSASTSEESENENP
HARGSFSYKSLRKGGPSQREQYLPGAIALFNVNNSSNKDQEPEEKKKKKKEKKSKSDDKN ENKNDPEKKKKKKDKEKKKKEEKSKDKKEEEKKEVVVIDPSGNTYYNWLFCITLPVMYNW TMVIARACFDELQSDYLEYWLILDYVSDIVYLIDMFVRTRTGYLEQGLLVKEELKLINKY KSNLQFKLDVLSLIPTDLLYFKLGWNYPEIRLNRLLRFSRMFEFFQRTETRTNYPNIFRI SNLVMYIVIIIHWNACVFYSISKAIGFGNDTWVYPDINDPEFGRLARKYVYSLYWSTLTL TTIGETPPPVRDSEYVFVVVDFLIGVLIFATIVGNIGSMISNMNAARAEFQARIDAIKQY MHFRNVSKDMEKRVIKWFDYLWTNKKTVDEKEVLKYLPDKLRAEIAINVHLDTLKKVRIF ADCEAGLLVELVLKLQPQVYSPGDYICKKGDIGREMYIIKEGKLAVVADDGVTQFVVLSD GSYFGEISILNIKGSKAGNRRTANIKSIGYSDLFCLSKDDLMEALTEYPDAKTMLEEKGK QILMKDGLLDLNIANAGSDPKDLEEKVTRMEGSVDLLQTRFARILAEYESMQQKLKQRLT KVEKFLKPLIDTEFSSIEGPGAESGPIDST Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 1 Clinical Trial Drugs | + | ||||
| 1 | dequalinium | Drug Info | Clinical trial | Myelopathy | [2] | |
| Mode of Action | [+] 1 Modes of Action | + | ||||
| Blocker (channel blocker) | [+] 3 Blocker (channel blocker) drugs | + | ||||
| 1 | dequalinium | Drug Info | [1] | |||
| 2 | H-8 | Drug Info | [3] | |||
| 3 | L-(cis)-diltiazem | Drug Info | [4] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Cholesterol | Ligand Info | |||||
| Structure Description | Cryo-EM structure of human Apo CNGA1 channel in K+/Ca2+ | PDB:7LFT | ||||
| Method | Electron microscopy | Resolution | 2.60 Å | Mutation | No | [5] |
| PDB Sequence |
VVIDPSGNTY
165 YNWLFCITLP175 VMYNWTMVIA185 RACFDELQSD195 YLEYWLILDY205 VSDIVYLIDM 215 FVRTRTGYLE225 QGLLVKEELK235 LINKYKSNLQ245 FKLDVLSLIP255 TDLLYFKLGW 265 NYPEIRLNRL275 LRFSRMFEFF285 QRTETRTNYP295 NIFRISNLVM305 YIVIIIHWNA 315 CVFYSISKAI325 GFGNDTWVYP335 DINDPEFGRL345 ARKYVYSLYW355 STLTLTTIGE 365 TPPPVRDSEY375 VFVVVDFLIG385 VLIFATIVGN395 IGSMISNMNA405 ARAEFQARID 415 AIKQYMHFRN425 VSKDMEKRVI435 KWFDYLWTNK445 KTVDEKEVLK455 YLPDKLRAEI 465 AINVHLDTLK475 KVRIFADCEA485 GLLVELVLKL495 QPQVYSPGDY505 ICKKGDIGRE 515 MYIIKEGKLA525 VVADDGVTQF535 VVLSDGSYFG545 EISILNIKGS555 KAGNRRTANI 565 KSIGYSDLFC575 LSKDDLMEAL585 TEYPDAKTML595 EEKGKQILMK605 |
|||||
|
|
||||||
| Ligand Name: Cyclic Guanosine Monophosphate | Ligand Info | |||||
| Structure Description | Cryo-EM structure of human cGMP-bound CNGA1_E365Q channel in Na+/Ca2+ | PDB:7LG1 | ||||
| Method | Electron microscopy | Resolution | 2.70 Å | Mutation | Yes | [5] |
| PDB Sequence |
VVIDPSGNTY
165 YNWLFCITLP175 VMYNWTMVIA185 RACFDELQSD195 YLEYWLILDY205 VSDIVYLIDM 215 FVRTRTGYLE225 QGLLVKEELK235 LINKYKSNLQ245 FKLDVLSLIP255 TDLLYFKLGW 265 NYPEIRLNRL275 LRFSRMFEFF285 QRTETRTNYP295 NIFRISNLVM305 YIVIIIHWNA 315 CVFYSISKAI325 GFGNDTWVYP335 DINDPEFGRL345 ARKYVYSLYW355 STLTLTTIGQ 365 TPPPVRDSEY375 VFVVVDFLIG385 VLIFATIVGN395 IGSMISNMNA405 ARAEFQARID 415 AIKQYMHFRN425 VSKDMEKRVI435 KWFDYLWTNK445 KTVDEKEVLK455 YLPDKLRAEI 465 AINVHLDTLK475 KVRIFADCEA485 GLLVELVLKL495 QPQVYSPGDY505 ICKKGDIGRE 515 MYIIKEGKLA525 VVADDGVTQF535 VVLSDGSYFG545 EISILNIKGS555 KAGNRRTANI 565 KSIGYSDLFC575 LSKDDLMEAL585 TEYPDAKTML595 EEKGKQILMK605 |
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
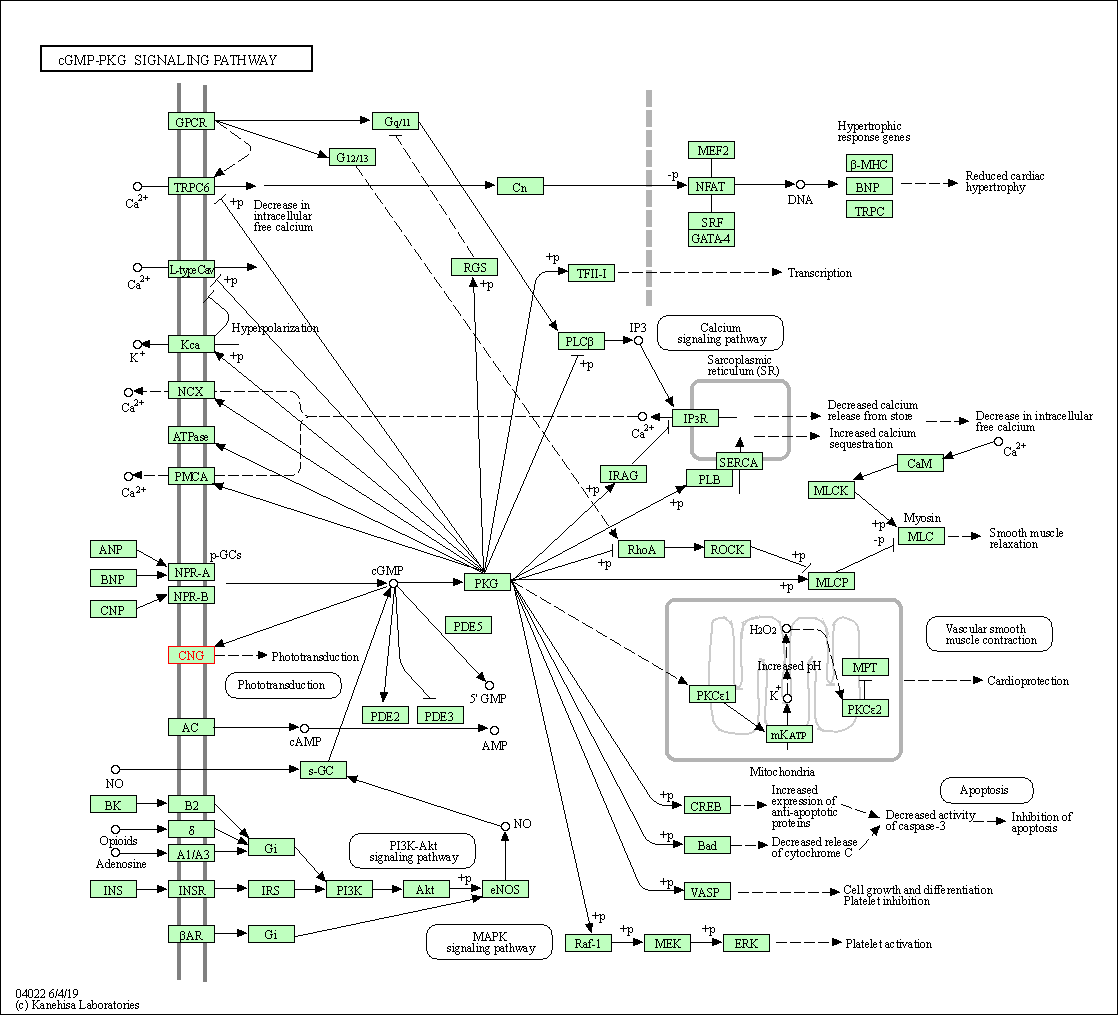
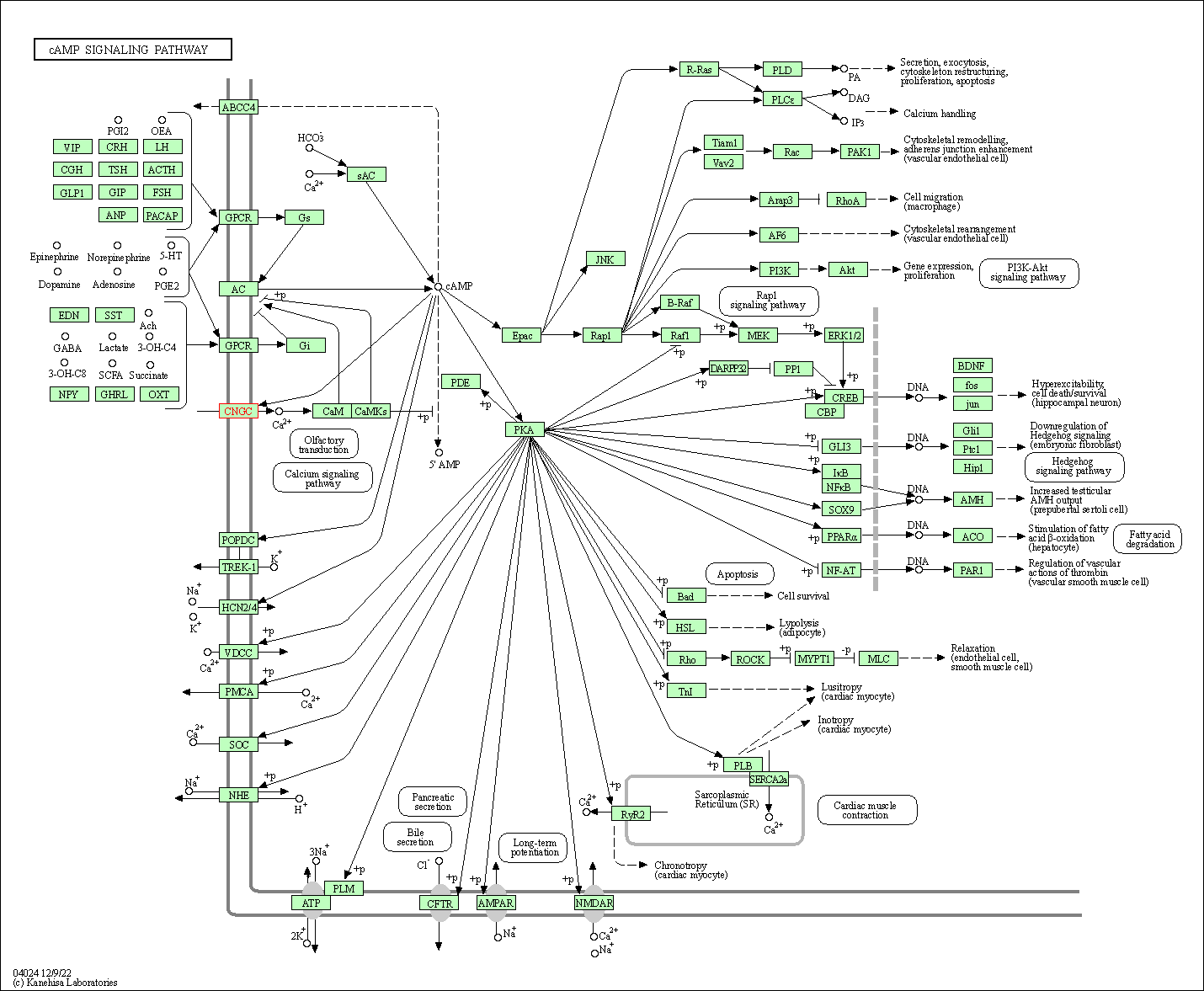
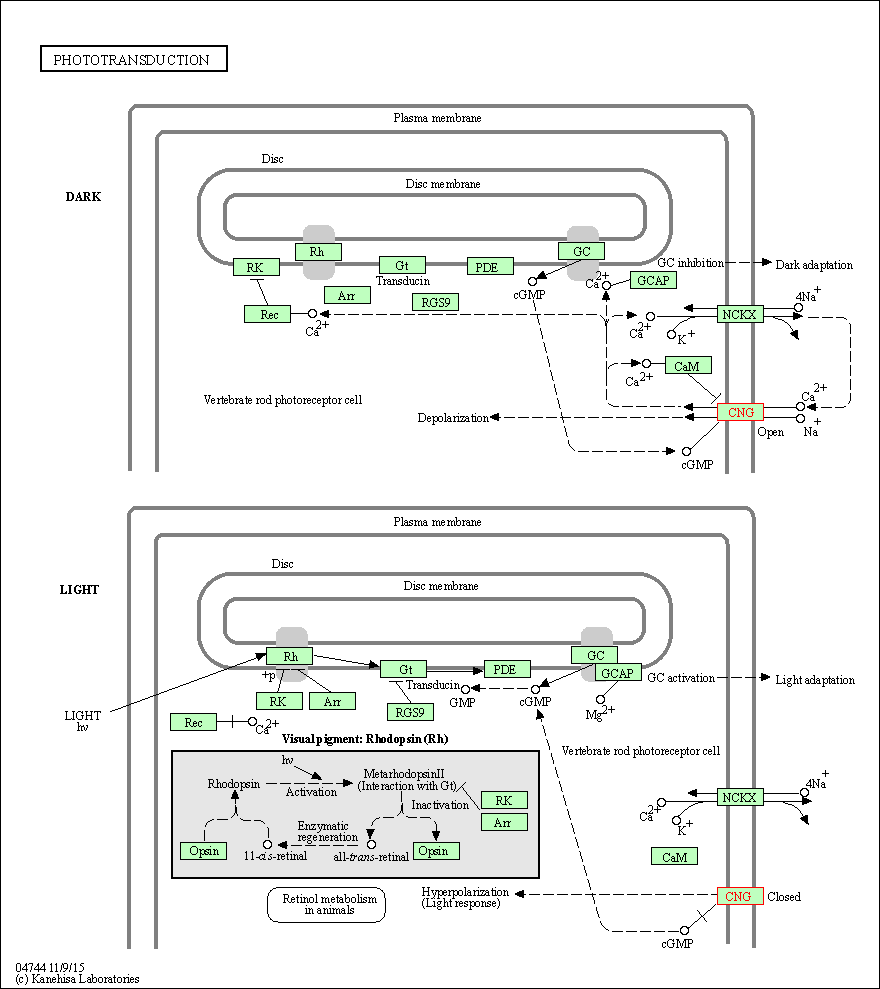
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| cGMP-PKG signaling pathway | hsa04022 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| cAMP signaling pathway | hsa04024 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Phototransduction | hsa04744 | Affiliated Target |

|
| Class: Organismal Systems => Sensory system | Pathway Hierarchy | ||
| Degree | 5 | Degree centrality | 5.37E-04 | Betweenness centrality | 0.00E+00 |
|---|---|---|---|---|---|
| Closeness centrality | 1.66E-01 | Radiality | 1.26E+01 | Clustering coefficient | 1.00E+00 |
| Neighborhood connectivity | 7.60E+00 | Topological coefficient | 5.07E-01 | Eccentricity | 14 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Dequalinium: a novel, high-affinity blocker of CNGA1 channels. J Gen Physiol. 2003 Jan;121(1):37-47. | |||||
| REF 2 | Pharmacological characterisation of the human small conductance calcium-activated potassium channel hSK3 reveals sensitivity to tricyclic antidepressants and antipsychotic phenothiazines. Neuropharmacology. 2001 May;40(6):772-83. | |||||
| REF 3 | Direct blockade of both cloned rat rod photoreceptor cyclic nucleotide-gated non-selective cation (CNG) channel alpha-subunit and native CNG channels from Xenopus rod outer segments by H-8, a non-specific cyclic nucleotide-dependent protein kinase inhibitor. Neurosci Lett. 1997 Sep 12;233(1):37-40. | |||||
| REF 4 | A new subunit of the cyclic nucleotide-gated cation channel in retinal rods. Nature. 1993 Apr 22;362(6422):764-7. | |||||
| REF 5 | Structural mechanisms of gating and selectivity of human rod CNGA1 channel. Neuron. 2021 Apr 21;109(8):1302-1313.e4. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

