Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T46642
(Former ID: TTDC00062)
|
|||||
| Target Name |
Arachidonate 5-lipoxygenase activating protein (FLAP)
|
|||||
| Synonyms |
MK-886-binding protein; FLAP; ALOX5AP
Click to Show/Hide
|
|||||
| Gene Name |
ALOX5AP
|
|||||
| Target Type |
Discontinued target
|
[1] | ||||
| Disease | [+] 4 Target-related Diseases | + | ||||
| 1 | Asthma [ICD-11: CA23] | |||||
| 2 | Cardiovascular disease [ICD-11: BA00-BE2Z] | |||||
| 3 | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||||
| 4 | Allergic/hypersensitivity disorder [ICD-11: 4A80-4A8Z] | |||||
| Function |
Required for leukotrienebiosynthesis by ALOX5 (5- lipoxygenase). Anchors ALOX5 to the membrane. Binds arachidonic acid, and could play an essential role in the transfer of arachidonic acid to ALOX5. Binds to MK-886, a compound that blocks the biosynthesis of leukotrienes.
Click to Show/Hide
|
|||||
| UniProt ID | ||||||
| Sequence |
MDQETVGNVVLLAIVTLISVVQNGFFAHKVEHESRTQNGRSFQRTGTLAFERVYTANQNC
VDAYPTFLAVLWSAGLLCSQVPAAFAGLMYLFVRQKYFVGYLGERTQSTPGYIFGKRIIL FLFLMSVAGIFNYYLIFFFGSDFENYIKTISTTISPLLLIP Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Discontinued Drug(s) | [+] 6 Discontinued Drugs | + | ||||
| 1 | DG031 | Drug Info | Discontinued in Phase 3 | Asthma | [2], [3] | |
| 2 | AM103 | Drug Info | Discontinued in Phase 2 | Cardiovascular disease | [4] | |
| 3 | GSK2190914 | Drug Info | Discontinued in Phase 2 | Asthma | [4] | |
| 4 | GSK2190915 | Drug Info | Discontinued in Phase 2 | Asthma | [5] | |
| 5 | AZD4769 | Drug Info | Discontinued in Phase 1 | Solid tumour/cancer | [6] | |
| 6 | A-93178 | Drug Info | Terminated | Allergy | [7] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Inhibitor | [+] 7 Inhibitor drugs | + | ||||
| 1 | DG031 | Drug Info | [1], [8] | |||
| 2 | AM103 | Drug Info | [9] | |||
| 3 | GSK2190914 | Drug Info | [3] | |||
| 4 | GSK2190915 | Drug Info | [10] | |||
| 5 | AZD4769 | Drug Info | [11] | |||
| 6 | L-671,480 | Drug Info | [12] | |||
| 7 | L-689,037 | Drug Info | [11] | |||
| Modulator | [+] 1 Modulator drugs | + | ||||
| 1 | A-93178 | Drug Info | [7] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: MK-591 | Ligand Info | |||||
| Structure Description | Crystal structure of human FLAP with MK-591 | PDB:2Q7M | ||||
| Method | X-ray diffraction | Resolution | 4.25 Å | Mutation | Yes | [13] |
| PDB Sequence |
MDQETVGNVV
10 LLAIVTLISV20 VQNGFFAHKV30 EHESRTQNGR40 SFQRTGTLAF50 ERVYTANQNC 60 VDAYPTFLAV70 LWSAGLLCSQ80 VPAAFAGLMY90 LFVRQKYFVG100 YLGERTQSTP 110 GYIFGKRIIL120 FLFLMSVAGI130 FNYYLIFFF
|
|||||
|
|
VAL20
4.503
VAL21
3.472
ASN23
3.310
GLY24
2.976
PHE25
2.881
ALA27
3.241
HIS28
3.652
ASN57
4.538
ASN59
4.362
VAL61
4.522
ASP62
3.235
|
|||||
| Ligand Name: DG031 | Ligand Info | |||||
| Structure Description | Crystal Structures of FLAP bound to DG-031 | PDB:6VGC | ||||
| Method | X-ray diffraction | Resolution | 2.37 Å | Mutation | No | [14] |
| PDB Sequence |
SLDQETVGNV
9 VLLAIVTLIS19 VVQNGFFAHK29 VEHESRTTGT47 LAFERVYTAN57 QNCVDAYPTF 67 LAVLWSAGLL77 CSQVPAAFAG87 LMYLFVRQKY97 FVGYLPGYIF114 GKRIILFLFL 124 MSVAGIFNYY134 LIFFFGSDFE144 NYIATISTTI154 SPLL
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Fc epsilon RI signaling pathway | hsa04664 | Affiliated Target |
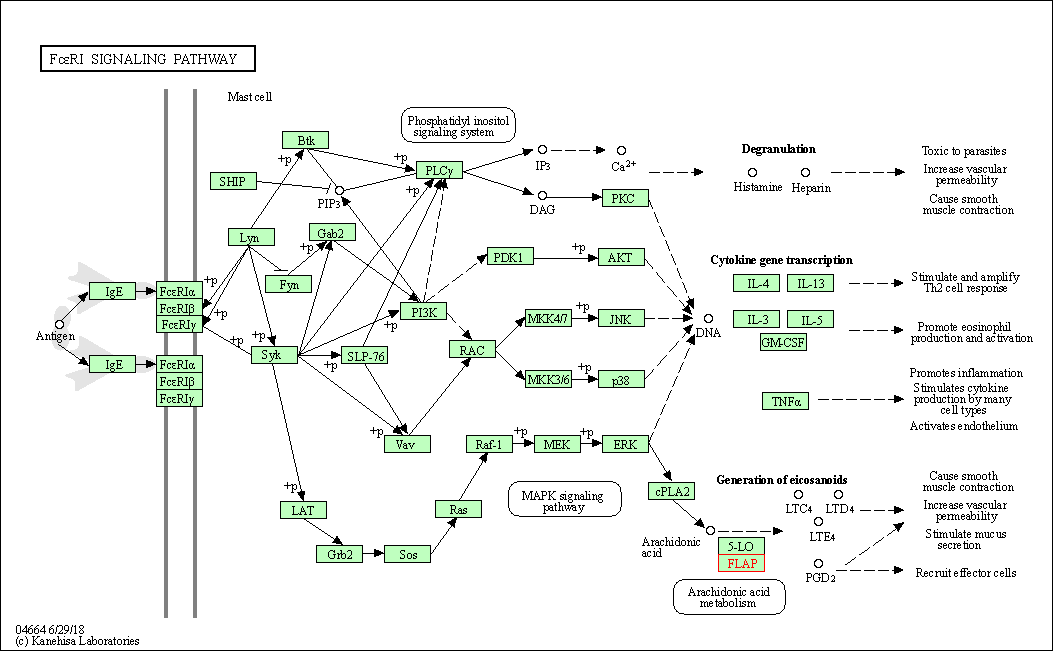
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Degree | 1 | Degree centrality | 1.07E-04 | Betweenness centrality | 0.00E+00 |
|---|---|---|---|---|---|
| Closeness centrality | 1.60E-01 | Radiality | 1.24E+01 | Clustering coefficient | 0.00E+00 |
| Neighborhood connectivity | 7.00E+00 | Topological coefficient | 1.00E+00 | Eccentricity | 14 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-interacting Proteins | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| NetPath Pathway | [+] 3 NetPath Pathways | + | ||||
| 1 | IL4 Signaling Pathway | |||||
| 2 | TGF_beta_Receptor Signaling Pathway | |||||
| 3 | Leptin Signaling Pathway | |||||
| Panther Pathway | [+] 1 Panther Pathways | + | ||||
| 1 | Inflammation mediated by chemokine and cytokine signaling pathway | |||||
| WikiPathways | [+] 4 WikiPathways | + | ||||
| 1 | Nuclear Receptors Meta-Pathway | |||||
| 2 | Arachidonic acid metabolism | |||||
| 3 | Eicosanoid Synthesis | |||||
| 4 | Selenium Micronutrient Network | |||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | BAY x 1005 attenuates atherosclerosis in apoE/LDLR - double knockout mice. J Physiol Pharmacol. 2007 Sep;58(3):583-8. | |||||
| REF 2 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 5148). | |||||
| REF 3 | FLAP inhibitors for the treatment of inflammatory diseases. Curr Opin Investig Drugs. 2009 Nov;10(11):1163-72. | |||||
| REF 4 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800026047) | |||||
| REF 5 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800027280) | |||||
| REF 6 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800024847) | |||||
| REF 7 | Characterization of A-93178, an iminoxy-quinoline inhibitor of leukotriene biosynthesis. Adv Exp Med Biol. 1997;433:91-4. | |||||
| REF 8 | Effects of a 5-lipoxygenase-activating protein inhibitor on biomarkers associated with risk of myocardial infarction: a randomized trial. JAMA. 2005 May 11;293(18):2245-56. | |||||
| REF 9 | AM103 Experimental Treatment for Respiratory Diseases. Amira Pharmaceuticals/GSK. 2009. | |||||
| REF 10 | Clinical pipeline report, company report or official report of GlaxoSmithKline (2009). | |||||
| REF 11 | 5-Lipoxygenase-activating protein is the target of a novel hybrid of two classes of leukotriene biosynthesis inhibitors. Mol Pharmacol. 1992 Feb;41(2):267-72. | |||||
| REF 12 | 5-Lipoxygenase-activating protein is the target of a quinoline class of leukotriene synthesis inhibitors. Mol Pharmacol. 1991 Jul;40(1):22-7. | |||||
| REF 13 | Crystal structure of inhibitor-bound human 5-lipoxygenase-activating protein. Science. 2007 Jul 27;317(5837):510-2. | |||||
| REF 14 | Structure-based, multi-targeted drug discovery approach to eicosanoid inhibition: Dual inhibitors of mPGES-1 and 5-lipoxygenase activating protein (FLAP). Biochim Biophys Acta Gen Subj. 2021 Feb;1865(2):129800. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

