Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T17448
(Former ID: TTDS00089)
|
|||||
| Target Name |
Histamine N-methyltransferase (HNMT)
|
|||||
| Synonyms |
Histamine-N-methyltransferase; HNMT; HMT
Click to Show/Hide
|
|||||
| Gene Name |
HNMT
|
|||||
| Target Type |
Successful target
|
[1] | ||||
| Disease | [+] 2 Target-related Diseases | + | ||||
| 1 | Episodic vestibular syndrome [ICD-11: AB31] | |||||
| 2 | Malaria [ICD-11: 1F40-1F45] | |||||
| Function |
Inactivates histamine by N-methylation. Plays an important role in degrading histamine and in regulating the airway response to histamine.
Click to Show/Hide
|
|||||
| BioChemical Class |
Methyltransferase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.1.1.8
|
|||||
| Sequence |
MASSMRSLFSDHGKYVESFRRFLNHSTEHQCMQEFMDKKLPGIIGRIGDTKSEIKILSIG
GGAGEIDLQILSKVQAQYPGVCINNEVVEPSAEQIAKYKELVAKTSNLENVKFAWHKETS SEYQSRMLEKKELQKWDFIHMIQMLYYVKDIPATLKFFHSLLGTNAKMLIIVVSGSSGWD KLWKKYGSRFPQDDLCQYITSDDLTQMLDNLGLKYECYDLLSTMDISDCFIDGNENGDLL WDFLTETCNFNATAPPDLRAELGKDLQEPEFSAKKEGKVLFNNTLSFIVIEA Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Approved Drug(s) | [+] 2 Approved Drugs | + | ||||
| 1 | Amodiaquine | Drug Info | Approved | Malaria | [2] | |
| 2 | Diphenhydramine | Drug Info | Approved | Meniere disease | [3] | |
| Clinical Trial Drug(s) | [+] 1 Clinical Trial Drugs | + | ||||
| 1 | Metoprine | Drug Info | Phase 2 | Advanced cancer | [4], [5] | |
| Mode of Action | [+] 1 Modes of Action | + | ||||
| Inhibitor | [+] 5 Inhibitor drugs | + | ||||
| 1 | Amodiaquine | Drug Info | [1] | |||
| 2 | Diphenhydramine | Drug Info | [6] | |||
| 3 | Metoprine | Drug Info | [6] | |||
| 4 | (L-)-S-adenosyl-L-homocysteine | Drug Info | [7] | |||
| 5 | 4-(DIMETHYLAMINO)BUTYL IMIDOTHIOCARBAMATE | Drug Info | [6] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Diphenhydramine | Ligand Info | |||||
| Structure Description | Histamine Methyltransferase Complexed with the Antihistamine Drug Diphenhydramine | PDB:2AOT | ||||
| Method | X-ray diffraction | Resolution | 1.90 Å | Mutation | Yes | [8] |
| PDB Sequence |
> Chain A
MRSLFSDHGK 14 YVESFRRFLN24 HSTEHQCMQE34 FMDKKLPGII44 GRIGDTKSEI54 KILSIGGGAG 64 EIDLQILSKV74 QAQYPGVCIN84 NEVVEPSAEQ94 IAKYKELVAK104 TSNLENVKFA 114 WHKETSSEYQ124 SRMLEKKELQ134 KWDFIHMIQM144 LYYVKDIPAT154 LKFFHSLLGT 164 NAKMLIIVVS174 GSSGWDKLWK184 KYGSRFPQDD194 LCQYITSDDL204 TQMLDNLGLK 214 YEYDLLSTMD225 ISDCFIDGNE235 NGDLLWDFLT245 ETNFNATAPP256 DLRAELGKDL 266 QEPEFSAKKE276 GKVLFNNTLS286 FIVIEA> Chain B MRSLFSDHGK 14 YVESFRRFLN24 HSTEHQCMQE34 FMDKKLPGII44 GRIGDTKSEI54 KILSIGGGAG 64 EIDLQILSKV74 QAQYPGVINN85 EVVEPSAEQI95 AKYKELVAKT105 SNLENVKFAW 115 HKETSSEYQS125 RMLEKKELQK135 WDFIHMIQML145 YYVKDIPATL155 KFFHSLLGTN 165 AKMLIIVVSG175 SSGWDKLWKK185 YGSRFPQDDL195 CQYITSDDLT205 QMLDNLGLKY 215 ECYDLLSTMD225 ISDCFIDGNE235 NGDLLWDFLT245 ETNFNATAPP256 DLRAELGKDL 266 QEPEFSAKKE276 GKVLFNNTLS286 FIVIEA
|
|||||
|
|
LEU8[A]
4.476
PHE9[A]
3.923
PHE19[A]
4.015
PHE22[A]
3.718
GLU28[A]
4.600
GLN143[A]
3.702
TYR146[A]
3.522
TYR147[A]
3.516
VAL173[A]
3.892
TRP179[A]
3.598
TRP183[A]
3.539
CYS196[A]
3.747
GLN197[A]
3.575
TYR198[A]
3.466
PHE243[A]
3.470
GLU246[A]
4.369
ASN283[A]
4.731
LEU8[B]
4.123
PHE9[B]
3.722
TYR15[B]
3.406
PHE19[B]
4.371
PHE22[B]
3.602
GLU28[B]
4.958
GLN143[B]
3.905
TYR146[B]
3.530
TYR147[B]
3.589
VAL173[B]
3.909
TRP179[B]
3.473
TRP183[B]
3.550
CYS196[B]
3.160
GLN197[B]
3.423
TYR198[B]
3.320
ILE199[B]
4.885
PHE243[B]
3.485
GLU246[B]
3.108
ASN283[B]
4.674
|
|||||
| Ligand Name: Ergotidine | Ligand Info | |||||
| Structure Description | Crystal Structure Analysis of Human Histamine Methyltransferase (Thr105 Polymorphic Variant) Complexed with AdoHcy and Histamine | PDB:1JQD | ||||
| Method | X-ray diffraction | Resolution | 2.28 Å | Mutation | No | [9] |
| PDB Sequence |
MRSLFHGKYV
16 ESFRRFLNHS26 TEHQCMQEFM36 DKKLPGIIGR46 IGDTKSEIKI56 LSIGGGAGEI 66 DLQILSKVQA76 QYPGVCINNE86 VVEPSAEQIA96 KYKELVAKTS106 NLENVKFAWH 116 KETSSEYQSR126 MLEKKELQKW136 DFIHMIQMLY146 YVKDIPATLK156 FFHSLLGTNA 166 KMLIIVVSGS176 SGWDKLWKKY186 GSRFPQDDLC196 QYITSDDLTQ206 MLDNLGLKYE 216 CYDLLSTMDI226 SDCFIDGNEN236 GDLLWDFLTE246 TCNFNATAPP256 DLRAELGKDL 266 QEPEFSAKKE276 GKVLFNNTLS286 FIVIEA
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
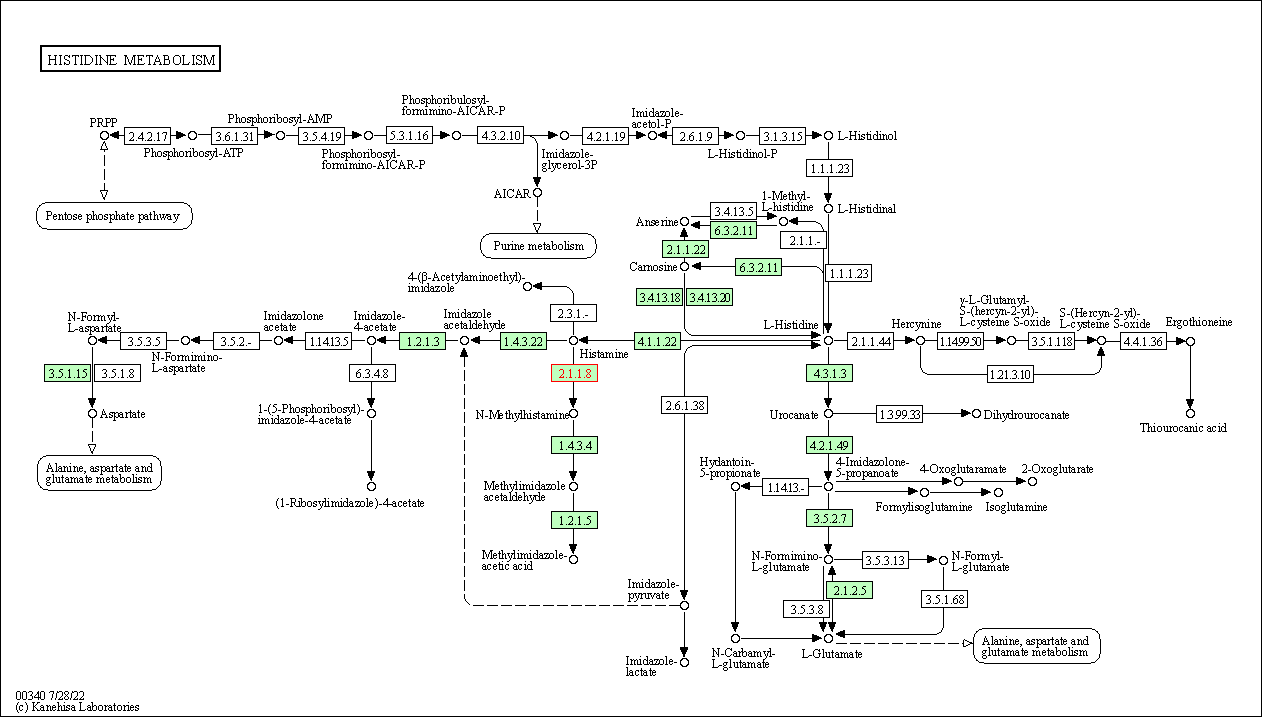
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Histidine metabolism | hsa00340 | Affiliated Target |

|
| Class: Metabolism => Amino acid metabolism | Pathway Hierarchy | ||
| Degree | 3 | Degree centrality | 3.22E-04 | Betweenness centrality | 4.04E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 1.94E-01 | Radiality | 1.33E+01 | Clustering coefficient | 3.33E-01 |
| Neighborhood connectivity | 2.13E+01 | Topological coefficient | 3.44E-01 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| BioCyc | [+] 1 BioCyc Pathways | + | ||||
| 1 | Histamine degradation | |||||
| KEGG Pathway | [+] 1 KEGG Pathways | + | ||||
| 1 | Histidine metabolism | |||||
| NetPath Pathway | [+] 1 NetPath Pathways | + | ||||
| 1 | TGF_beta_Receptor Signaling Pathway | |||||
| Pathwhiz Pathway | [+] 1 Pathwhiz Pathways | + | ||||
| 1 | Histidine Metabolism | |||||
| WikiPathways | [+] 2 WikiPathways | + | ||||
| 1 | Methylation Pathways | |||||
| 2 | Metapathway biotransformation | |||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Effect of amodiaquine, a histamine N-methyltransferase inhibitor, on, Propionibacterium acnes and lipopolysaccharide-induced hepatitis in mice. Eur J Pharmacol. 2007 Mar 8;558(1-3):179-84. | |||||
| REF 2 | Drug information of Amodiaquine, 2008. eduDrugs. | |||||
| REF 3 | Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services. 2015 | |||||
| REF 4 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7412). | |||||
| REF 5 | ClinicalTrials.gov (NCT01579110) Efficacy and Safety of Levamisole Combined With Standard Prednisolone in Warm Antibody Autoimmune Hemolytic Anemia.. U.S. National Institutes of Health. | |||||
| REF 6 | The Protein Data Bank. Nucleic Acids Res. 2000 Jan 1;28(1):235-42. | |||||
| REF 7 | How many drug targets are there Nat Rev Drug Discov. 2006 Dec;5(12):993-6. | |||||
| REF 8 | Structural basis for inhibition of histamine N-methyltransferase by diverse drugs. J Mol Biol. 2005 Oct 21;353(2):334-344. | |||||
| REF 9 | Two polymorphic forms of human histamine methyltransferase: structural, thermal, and kinetic comparisons. Structure. 2001 Sep;9(9):837-49. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

