Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T14731
(Former ID: TTDC00052)
|
|||||
| Target Name |
NAD-dependent deacetylase sirtuin-1 (SIRT1)
|
|||||
| Synonyms |
hSIRT1; hSIR2; SIR2L1; SIR2-like protein 1; Regulatory protein SIR2 homolog 1; NAD-dependent protein deacetylase sirtuin-1
Click to Show/Hide
|
|||||
| Gene Name |
SIRT1
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 3 Target-related Diseases | + | ||||
| 1 | Rheumatoid arthritis [ICD-11: FA20] | |||||
| 2 | Systemic sclerosis [ICD-11: 4A42] | |||||
| 3 | Vasculitis [ICD-11: 4A44] | |||||
| Function |
Can modulate chromatin function through deacetylation of histones and can promote alterations in the methylation of histones and DNA, leading to transcriptional repression. Deacetylates a broad range of transcription factors and coregulators, thereby regulating target gene expression positively and negatively. Serves as a sensor of the cytosolic ratio of NAD(+)/NADH which is altered by glucose deprivation and metabolic changes associated with caloric restriction. Is essential in skeletal muscle cell differentiation and in response to low nutrients mediates the inhibitory effect on skeletal myoblast differentiation which also involves 5'-AMP-activated protein kinase (AMPK) and nicotinamide phosphoribosyltransferase (NAMPT). Component of the eNoSC (energy-dependent nucleolar silencing) complex, a complex that mediates silencing of rDNA in response to intracellular energy status and acts by recruiting histone-modifying enzymes. The eNoSC complex is able to sense the energy status of cell: upon glucose starvation, elevation of NAD(+)/NADP(+) ratio activates SIRT1, leading to histone H3 deacetylation followed by dimethylation of H3 at 'Lys-9' (H3K9me2) by SUV39H1 and the formation of silent chromatin in the rDNA locus. Deacetylates 'Lys-266' of SUV39H1, leading to its activation. Inhibits skeletal muscle differentiation by deacetylating PCAF and MYOD1. Deacetylates H2A and 'Lys-26' of HIST1H1E. Deacetylates 'Lys-16' of histone H4 (in vitro). Involved in NR0B2/SHP corepression function through chromatin remodeling: Recruited to LRH1 target gene promoters by NR0B2/SHP thereby stimulating histone H3 and H4 deacetylation leading to transcriptional repression. Proposed to contribute to genomic integrity via positive regulation of telomere length; however, reports on localization to pericentromeric heterochromatin are conflicting. Proposed to play a role in constitutive heterochromatin (CH) formation and/or maintenance through regulation of the available pool of nuclear SUV39H1. Upon oxidative/metabolic stress decreases SUV39H1 degradation by inhibiting SUV39H1 polyubiquitination by MDM2. This increase in SUV39H1 levels enhances SUV39H1 turnover in CH, which in turn seems to accelerate renewal of the heterochromatin which correlates with greater genomic integrity during stress response. Deacetylates 'Lys-382' of p53/TP53 and impairs its ability to induce transcription-dependent proapoptotic program and modulate cell senescence. Deacetylates TAF1B and thereby represses rDNA transcription by the RNA polymerase I. Deacetylates MYC, promotes the association of MYC with MAX and decreases MYC stability leading to compromised transformational capability. Deacetylates FOXO3 in response to oxidative stress thereby increasing its ability to induce cell cycle arrest and resistance to oxidative stress but inhibiting FOXO3-mediated induction of apoptosis transcriptional activity; also leading to FOXO3 ubiquitination and protesomal degradation. Appears to have a similar effect on MLLT7/FOXO4 in regulation of transcriptional activity and apoptosis. Deacetylates DNMT1; thereby impairs DNMT1 methyltransferase-independent transcription repressor activity, modulates DNMT1 cell cycle regulatory function and DNMT1-mediated gene silencing. Deacetylates RELA/NF-kappa-B p65 thereby inhibiting its transactivating potential and augments apoptosis in response to TNF-alpha. Deacetylates HIF1A, KAT5/TIP60, RB1 and HIC1. Deacetylates FOXO1 resulting in its nuclear retention and enhancement of its transcriptional activity leading to increased gluconeogenesis in liver. Inhibits E2F1 transcriptional activity and apoptotic function, possibly by deacetylation. Involved in HES1- and HEY2-mediated transcriptional repression. In cooperation with MYCN seems to be involved in transcriptional repression of DUSP6/MAPK3 leading to MYCN stabilization by phosphorylation at 'Ser-62'. Deacetylates MEF2D. Required for antagonist-mediated transcription suppression of AR-dependent genes which may be linked to local deacetylation of histone H3. Represses HNF1A-mediated transcription. Required for the repression of ESRRG by CREBZF. Deacetylates NR1H3 and NR1H2 and deacetylation of NR1H3 at 'Lys-434' positively regulates transcription of NR1H3:RXR target genes, promotes NR1H3 proteosomal degradation and results in cholesterol efflux; a promoter clearing mechanism after reach round of transcription is proposed. Involved in lipid metabolism. Implicated in regulation of adipogenesis and fat mobilization in white adipocytes by repression of PPARG which probably involves association with NCOR1 and SMRT/NCOR2. Deacetylates p300/EP300 and PRMT1. Deacetylates ACSS2 leading to its activation, and HMGCS1 deacetylation. Involved in liver and muscle metabolism. Through deacteylation and activation of PPARGC1A is required to activate fatty acid oxidation in skeletel muscle under low-glucose conditions and is involved in glucose homeostasis. Involved in regulation of PPARA and fatty acid beta-oxidation in liver. Involved in positive regulation of insulin secretion in pancreatic beta cells in response to glucose; the function seems to imply transcriptional repression of UCP2. Proposed to deacetylate IRS2 thereby facilitating its insulin-induced tyrosine phosphorylation. Deacetylates SREBF1 isoform SREBP-1C thereby decreasing its stability and transactivation in lipogenic gene expression. Involved in DNA damage response by repressing genes which are involved in DNA repair, such as XPC and TP73, deacetylating XRCC6/Ku70, and faciliting recruitment of additional factors to sites of damaged DNA, such as SIRT1-deacetylated NBN can recruit ATM to initiate DNA repair and SIRT1-deacetylated XPA interacts with RPA2. Also involved in DNA repair of DNA double-strand breaks by homologous recombination and specifically single-strand annealing independently of XRCC6/Ku70 and NBN. Transcriptional suppression of XPC probably involves an E2F4:RBL2 suppressor complex and protein kinase B (AKT) signaling. Transcriptional suppression of TP73 probably involves E2F4 and PCAF. Deacetylates WRN thereby regulating its helicase and exonuclease activities and regulates WRN nuclear translocation in response to DNA damage. Deacetylates APEX1 at 'Lys-6' and 'Lys-7' and stimulates cellular AP endonuclease activity by promoting the association of APEX1 to XRCC1. Increases p53/TP53-mediated transcription-independent apoptosis by blocking nuclear translocation of cytoplasmic p53/TP53 and probably redirecting it to mitochondria. Deacetylates XRCC6/Ku70 at 'Lys-539' and 'Lys-542' causing it to sequester BAX away from mitochondria thereby inhibiting stress-induced apoptosis. Is involved in autophagy, presumably by deacetylating ATG5, ATG7 and MAP1LC3B/ATG8. Deacetylates AKT1 which leads to enhanced binding of AKT1 and PDK1 to PIP3 and promotes their activation. Proposed to play role in regulation of STK11/LBK1-dependent AMPK signaling pathways implicated in cellular senescence which seems to involve the regulation of the acetylation status of STK11/LBK1. Can deacetylate STK11/LBK1 and thereby increase its activity, cytoplasmic localization and association with STRAD; however, the relevance of such activity in normal cells is unclear. In endothelial cells is shown to inhibit STK11/LBK1 activity and to promote its degradation. Deacetylates SMAD7 at 'Lys-64' and 'Lys-70' thereby promoting its degradation. Deacetylates CIITA and augments its MHC class II transactivation and contributes to its stability. Deacteylates MECOM/EVI1. Deacetylates PML at 'Lys-487' and this deacetylation promotes PML control of PER2 nuclear localization. During the neurogenic transition, repress selective NOTCH1-target genes through histone deacetylation in a BCL6-dependent manner and leading to neuronal differentiation. Regulates the circadian expression of several core clock genes, including ARNTL/BMAL1, RORC, PER2 and CRY1 and plays a critical role in maintaining a controlled rhythmicity in histone acetylation, thereby contributing to circadian chromatin remodeling. Deacetylates ARNTL/BMAL1 and histones at the circadian gene promoters in order to facilitate repression by inhibitory components of the circadian oscillator. Deacetylates PER2, facilitating its ubiquitination and degradation by the proteosome. Protects cardiomyocytes against palmitate-induced apoptosis. Deacetylates XBP1 isoform 2; deacetylation decreases protein stability of XBP1 isoform 2 and inhibits its transcriptional activity. Deacetylates PCK1 and directs its activity toward phosphoenolpyruvate production promoting gluconeogenesis. Involved in the CCAR2-mediated regulation of PCK1 and NR1D1. Deacetylates CTNB1 at 'Lys-49'. In POMC (pro-opiomelanocortin) neurons, required for leptin-induced activation of PI3K signaling. NAD-dependent protein deacetylase that links transcriptional regulation directly to intracellular energetics and participates in the coordination of several separated cellular functions such as cell cycle, response to DNA damage, metobolism, apoptosis and autophagy.
Click to Show/Hide
|
|||||
| BioChemical Class |
Carbon-nitrogen hydrolase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 3.5.1.-
|
|||||
| Sequence |
MADEAALALQPGGSPSAAGADREAASSPAGEPLRKRPRRDGPGLERSPGEPGGAAPEREV
PAAARGCPGAAAAALWREAEAEAAAAGGEQEAQATAAAGEGDNGPGLQGPSREPPLADNL YDEDDDDEGEEEEEAAAAAIGYRDNLLFGDEIITNGFHSCESDEEDRASHASSSDWTPRP RIGPYTFVQQHLMIGTDPRTILKDLLPETIPPPELDDMTLWQIVINILSEPPKRKKRKDI NTIEDAVKLLQECKKIIVLTGAGVSVSCGIPDFRSRDGIYARLAVDFPDLPDPQAMFDIE YFRKDPRPFFKFAKEIYPGQFQPSLCHKFIALSDKEGKLLRNYTQNIDTLEQVAGIQRII QCHGSFATASCLICKYKVDCEAVRGDIFNQVVPRCPRCPADEPLAIMKPEIVFFGENLPE QFHRAMKYDKDEVDLLIVIGSSLKVRPVALIPSSIPHEVPQILINREPLPHLHFDVELLG DCDVIINELCHRLGGEYAKLCCNPVKLSEITEKPPRTQKELAYLSELPPTPLHVSEDSSS PERTSPPDSSVIVTLLDQAAKSNDDLDVSESKGCMEEKPQEVQTSRNVESIAEQMENPDL KNVGSSTGEKNERTSVAGTVRKCWPNRVAKEQISRRLDGNQYLFLPPNRYIFHGAEVYSD SEDDVLSSSSCGSNSDSGTCQSPSLEEPMEDESEIEEFYNGLEDEPDVPERAGGAGFGTD GDDQEAINEAISVKQEVTDMNYPSNKS Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| ADReCS ID | BADD_A03103 ; BADD_A05675 | |||||
| HIT2.0 ID | T93SFQ | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 6 Clinical Trial Drugs | + | ||||
| 1 | Resveratrol | Drug Info | Phase 3 | Giant cell arteritis | [2] | |
| 2 | GSK2245840 | Drug Info | Phase 2 | Chronic obstructive pulmonary disease | [3] | |
| 3 | MB-12066 | Drug Info | Phase 2 | Obesity | [4] | |
| 4 | SEN-196 | Drug Info | Phase 2 | Huntington disease | [5], [6] | |
| 5 | SRT2379 | Drug Info | Phase 1 | Type-2 diabetes | [7] | |
| 6 | SRT3025 | Drug Info | Phase 1 | Type-2 diabetes | [8] | |
| Discontinued Drug(s) | [+] 1 Discontinued Drugs | + | ||||
| 1 | GSK184072 | Drug Info | Discontinued in Phase 2 | Type-2 diabetes | [9] | |
| Mode of Action | [+] 3 Modes of Action | + | ||||
| Inhibitor | [+] 16 Inhibitor drugs | + | ||||
| 1 | Resveratrol | Drug Info | [10] | |||
| 2 | SEN-196 | Drug Info | [1], [10] | |||
| 3 | CAMBINOL | Drug Info | [10], [16] | |||
| 4 | PMID25435179-Compound-WO2012106509CAY10602 | Drug Info | [10] | |||
| 5 | PMID25435179-Compound-WO2012106509Salermide | Drug Info | [10] | |||
| 6 | PMID25435179-Compound-WO2012106509Tenovin-6 | Drug Info | [10] | |||
| 7 | (R)-sirtinol | Drug Info | [18] | |||
| 8 | (S)-sirtinol | Drug Info | [18] | |||
| 9 | 2,3,4,9-tetrahydro-1H-carbazole-1-carboxamide | Drug Info | [19] | |||
| 10 | 2H-chromeno[2,3-d]pyrimidine-2,4(3H)-dione | Drug Info | [20] | |||
| 11 | Meta-sirtinol | Drug Info | [18] | |||
| 12 | Para-sirtinol | Drug Info | [18] | |||
| 13 | RO-316233 | Drug Info | [21] | |||
| 14 | Ro31-8220 | Drug Info | [21] | |||
| 15 | splitomicin | Drug Info | [20] | |||
| 16 | YK-3237 | Drug Info | [23] | |||
| Modulator | [+] 4 Modulator drugs | + | ||||
| 1 | GSK2245840 | Drug Info | [11], [12] | |||
| 2 | MB-12066 | Drug Info | [13] | |||
| 3 | SRT2379 | Drug Info | [14] | |||
| 4 | SRT3025 | Drug Info | [15] | |||
| Activator | [+] 2 Activator drugs | + | ||||
| 1 | GSK184072 | Drug Info | [17] | |||
| 2 | SRT1720 | Drug Info | [22] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Resveratrol | Ligand Info | |||||
| Structure Description | Crystal structure of SIRT1 in complex with resveratrol and an AMC-containing peptide | PDB:5BTR | ||||
| Method | X-ray diffraction | Resolution | 3.20 Å | Mutation | Yes | [24] |
| PDB Sequence |
DNLLFGDEII
153 TNGSDWTPRP180 RIGPYTFVQQ190 HLMIGTDPRT200 ILKDLLPETI210 PPPELDDMTL 220 WQIVINILSE230 PPKRKKRKDI240 NTIEDAVKLL250 QESKKIIVLT260 GAGVSVSSGI 270 PDFRSRDGIY280 ARLAVDFPDL290 PDPQAMFDIE300 YFRKDPRPFF310 KFAKEIYPGQ 320 FQPSLCHKFI330 ALSDKEGKLL340 RNYTQNIDTL350 EQVAGIQRII360 QCHGSFATAS 370 CLICKYKVDC380 EAVRGDIFNQ390 VVPRCPRCPA400 DEPLAIMKPE410 IVFFGENLPE 420 QFHRAMKYDK430 DEVDLLIVIG440 SSLKVRPVAL450 IPSSIPHEVP460 QILINREPLP 470 HLHFDVELLG480 DCDVIINELC490 HRLGGEYAKL500 SSNPVKLSEI510 TEQYLFLPPN 648 RYIFHGAEVY658
|
|||||
|
|
THR209
3.360
PRO212
3.605
GLU214
4.582
PRO291
4.328
ASP292
2.270
GLN294
3.329
ALA295
3.460
ASP298
2.260
PHE414
3.593
LYS444
3.303
VAL445
3.906
LEU202
3.712
LEU206
3.637
THR209
4.689
ILE210
3.940
PRO211
3.149
PRO212
3.973
|
|||||
| Ligand Name: Norleucine | Ligand Info | |||||
| Structure Description | SIRT1/Activator/Substrate Complex | PDB:4ZZJ | ||||
| Method | X-ray diffraction | Resolution | 2.74 Å | Mutation | No | [25] |
| PDB Sequence |
GPYTFVQQHL
192 MIGTDPRTIL202 KDLLPETIPP212 PELDDMTLWQ222 IVINILSEPP232 KRKKRKDINT 242 IEDAVKLLQE252 CKKIIVLTGA262 GVSVSCGIPD272 FRSRDGIYAR282 LAVDFPDLPD 292 PQAMFDIEYF302 RKDPRPFFKF312 AKEIYPGQFQ322 PSLCHKFIAL332 SDKEGKLLRN 342 YTQNIDTLEQ352 VAGIQRIIQC362 HGSFATASCL372 ICKYKVDCEA382 VRGDIFNQVV 392 PRCPRCPADE402 PLAIMKPEIV412 FFGENLPEQF422 HRAMKYDKDE432 VDLLIVIGSS 442 LKVRPVALIP452 SSIPHEVPQI462 LINREPLPHL472 HFDVELLGDC482 DVIINELCHR 492 LGGEYAKLCC502 NGGSQYLFLP646 PNRYIFHGAE656 VYSD
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Nicotinate and nicotinamide metabolism | hsa00760 | Affiliated Target |

|
| Class: Metabolism => Metabolism of cofactors and vitamins | Pathway Hierarchy | ||
| FoxO signaling pathway | hsa04068 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| AMPK signaling pathway | hsa04152 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
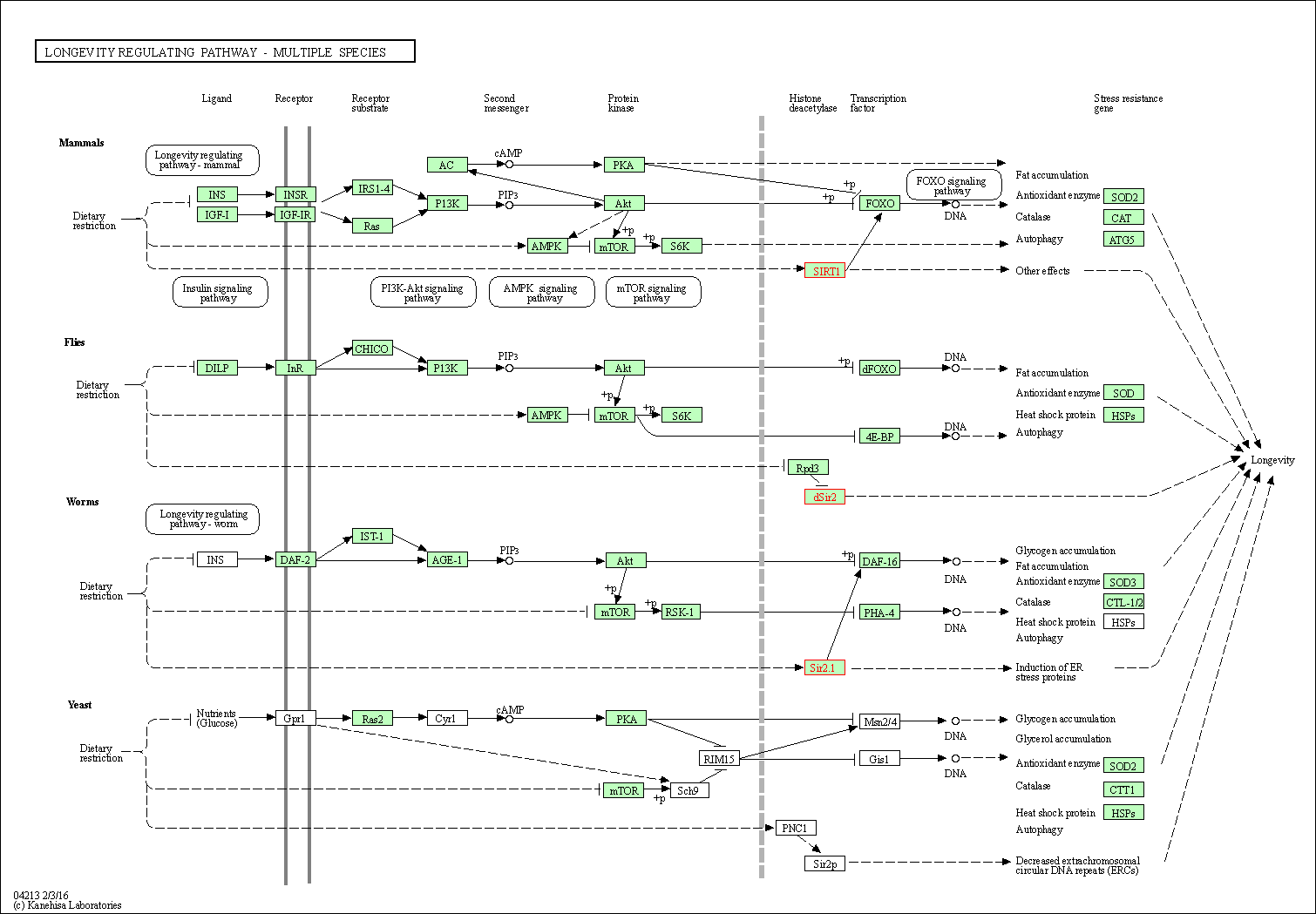
| Longevity regulating pathway | hsa04211 | Affiliated Target |

|
| Class: Organismal Systems => Aging | Pathway Hierarchy | ||
| Longevity regulating pathway - multiple species | hsa04213 | Affiliated Target |

|
| Class: Organismal Systems => Aging | Pathway Hierarchy | ||
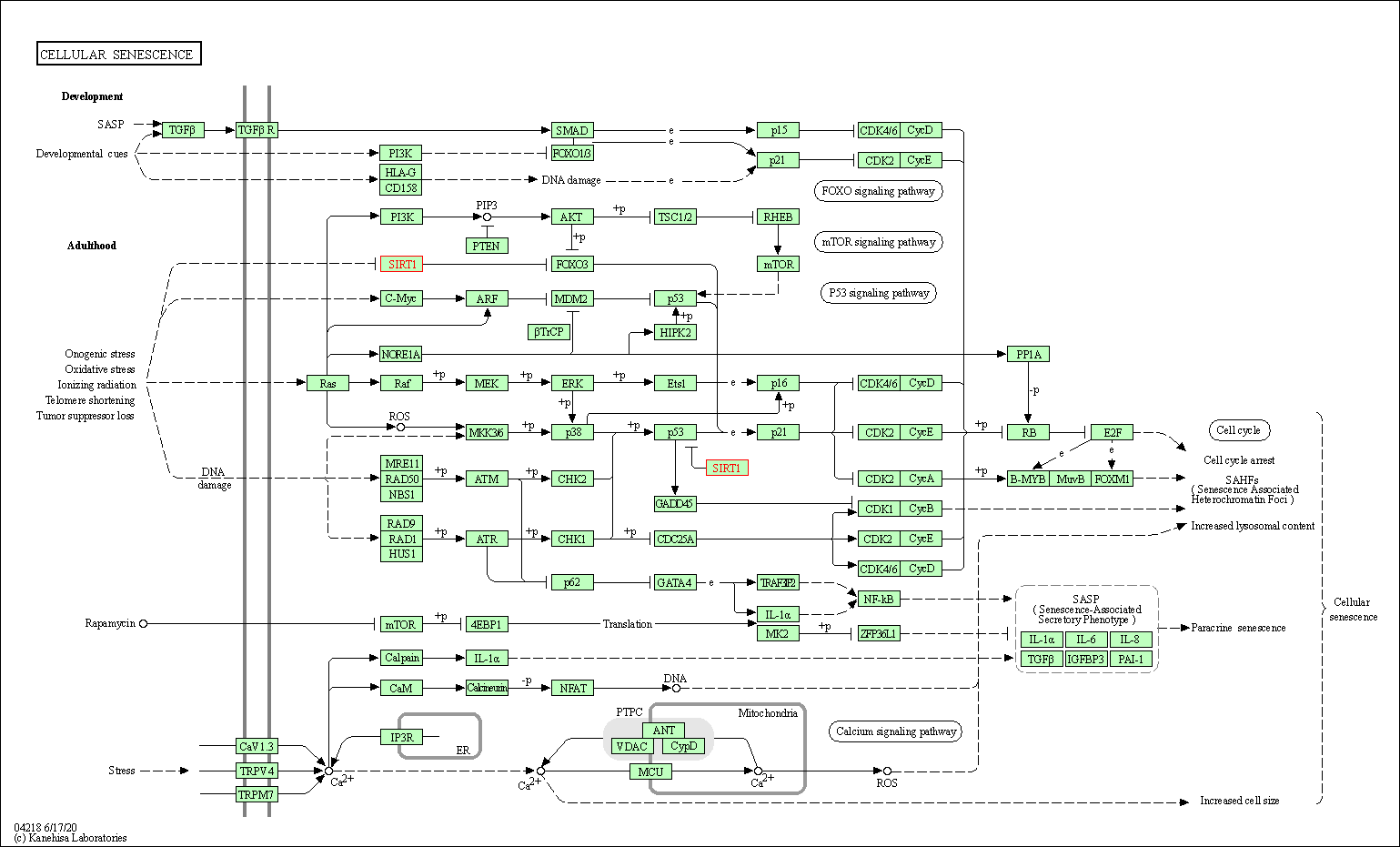
| Cellular senescence | hsa04218 | Affiliated Target |

|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
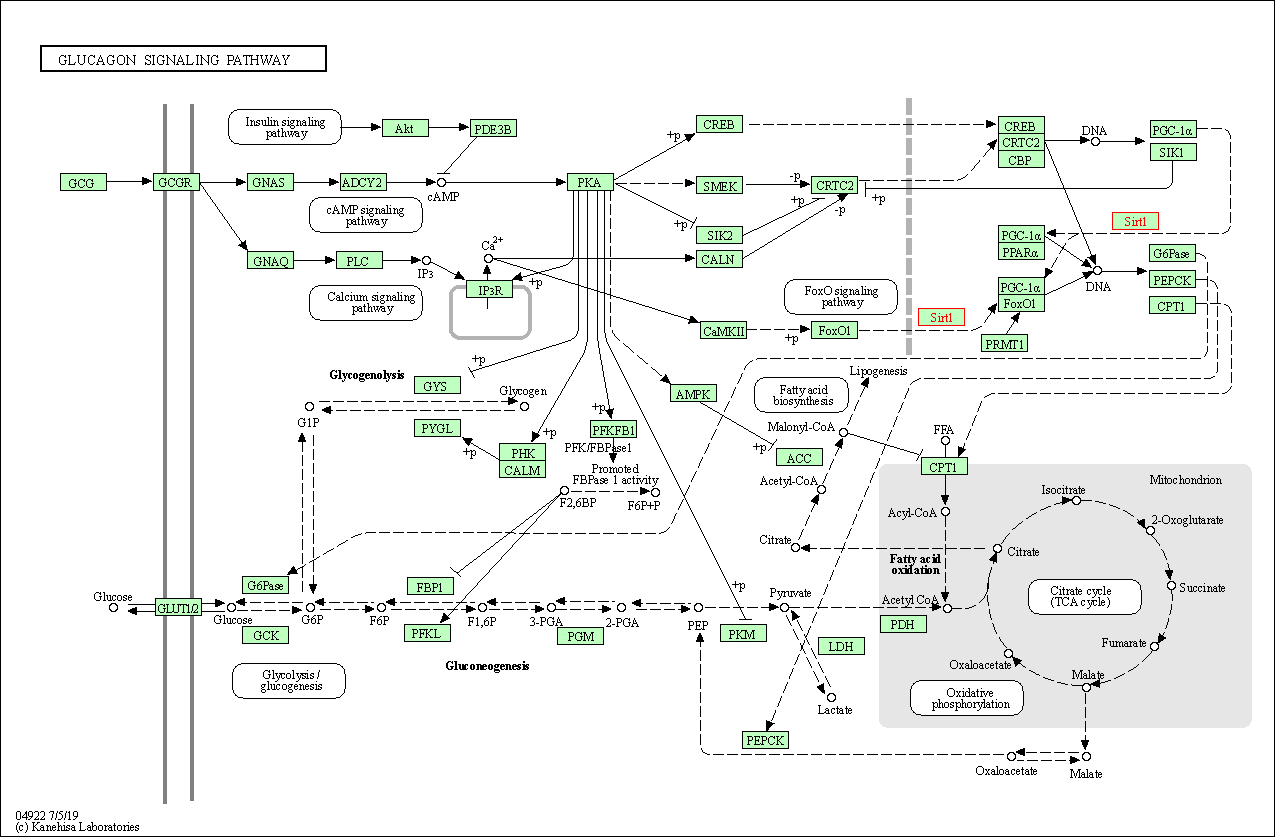
| Glucagon signaling pathway | hsa04922 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 46 | Degree centrality | 4.94E-03 | Betweenness centrality | 4.46E-03 |
|---|---|---|---|---|---|
| Closeness centrality | 2.56E-01 | Radiality | 1.44E+01 | Clustering coefficient | 9.95E-02 |
| Neighborhood connectivity | 3.97E+01 | Topological coefficient | 4.28E-02 | Eccentricity | 10 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) |
||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 5 KEGG Pathways | + | ||||
| 1 | FoxO signaling pathway | |||||
| 2 | AMPK signaling pathway | |||||
| 3 | Glucagon signaling pathway | |||||
| 4 | Amphetamine addiction | |||||
| 5 | MicroRNAs in cancer | |||||
| Panther Pathway | [+] 1 Panther Pathways | + | ||||
| 1 | p53 pathway | |||||
| PID Pathway | [+] 8 PID Pathways | + | ||||
| 1 | p73 transcription factor network | |||||
| 2 | Signaling events mediated by HDAC Class III | |||||
| 3 | E2F transcription factor network | |||||
| 4 | HIF-2-alpha transcription factor network | |||||
| 5 | Signaling events mediated by HDAC Class I | |||||
| 6 | FoxO family signaling | |||||
| 7 | Regulation of Androgen receptor activity | |||||
| 8 | Regulation of retinoblastoma protein | |||||
| Reactome | [+] 3 Reactome Pathways | + | ||||
| 1 | RORA activates gene expression | |||||
| 2 | Regulation of HSF1-mediated heat shock response | |||||
| 3 | Circadian Clock | |||||
| WikiPathways | [+] 5 WikiPathways | + | ||||
| 1 | Integrated Pancreatic Cancer Pathway | |||||
| 2 | SREBF and miR33 in cholesterol and lipid homeostasis | |||||
| 3 | Integrated Breast Cancer Pathway | |||||
| 4 | SREBP signalling | |||||
| 5 | Androgen receptor signaling pathway | |||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Sirtuin 1 (SIRT1): the misunderstood HDAC. J Biomol Screen. 2011 Dec;16(10):1153-69. | |||||
| REF 2 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 3 | ClinicalTrials.gov (NCT01154101) Study of the Clinical Activity, Safety, and Tolerability of SRT2104 in Subjects With Moderate to Severe Plaque-Type Psoriasis. U.S. National Institutes of Health. | |||||
| REF 4 | ClinicalTrials.gov (NCT02029586) Therapeutic Exploratory Phase 2 Study to Evaluate the Safety and Efficacy of MB12066 in Patients With Nonalcoholic Fatty Liver Disease(NAFLD) Except Cirrhosis. U.S. National Institutes of Health. | |||||
| REF 5 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 8100). | |||||
| REF 6 | ClinicalTrials.gov (NCT01521585) A Phase II Safety and Tolerability Study With SEN0014196. U.S. National Institutes of Health. | |||||
| REF 7 | ClinicalTrials.gov (NCT01018628) A Clinical Study to Assess the Safety and Pharmacokinetics of SRT2379 in Normal Healthy Male Volunteers. U.S. National Institutes of Health. | |||||
| REF 8 | ClinicalTrials.gov (NCT01340911) A Study in Healthy Male Volunteers to Investigate Different Doses of a New Drug for the Treatment of Metabolic Diseases. U.S. National Institutes of Health. | |||||
| REF 9 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800024569) | |||||
| REF 10 | Sirtuin modulators: an updated patent review (2012 - 2014).Expert Opin Ther Pat. 2015 Jan;25(1):5-15. | |||||
| REF 11 | Sirtuin 1 activator SRT2104 protects Huntington's disease mice. Ann Clin Transl Neurol. 2014 Dec;1(12):1047-52. | |||||
| REF 12 | Anti-inflammatory agents to treat or prevent type 2 diabetes, metabolic syndrome and cardiovascular disease. Expert Opin Investig Drugs. 2015 Mar;24(3):283-307. | |||||
| REF 13 | Pharmacological activation of Sirt1 ameliorates polyglutamine-induced toxicity through the regulation of autophagy.PLoS One.2013 Jun 10;8(6):e64953. | |||||
| REF 14 | SRT2379, a small-molecule SIRT1 activator, fails to reduce cytokine release in a human endotoxemia model. Critical Care 2013, 17(Suppl 4):P8. | |||||
| REF 15 | The Sirt1 Activators SRT2183 and SRT3025 Inhibit RANKL-Induced Osteoclastogenesis in Bone Marrow-Derived Macrophages and Down-Regulate Sirt3 in Sirt1 Null Cells. PLoS One. 2015 Jul 30;10(7):e0134391. | |||||
| REF 16 | Novel cambinol analogs as sirtuin inhibitors: synthesis, biological evaluation, and rationalization of activity. J Med Chem. 2009 May 14;52(9):2673-82. | |||||
| REF 17 | Clinical pipeline report, company report or official report of GlaxoSmithKline (2009). | |||||
| REF 18 | Design, synthesis, and biological evaluation of sirtinol analogues as class III histone/protein deacetylase (Sirtuin) inhibitors. J Med Chem. 2005 Dec 1;48(24):7789-95. | |||||
| REF 19 | Discovery of indoles as potent and selective inhibitors of the deacetylase SIRT1. J Med Chem. 2005 Dec 15;48(25):8045-54. | |||||
| REF 20 | Characterization of sirtuin inhibitors in nematodes expressing a muscular dystrophy protein reveals muscle cell and behavioral protection by specif... J Med Chem. 2010 Feb 11;53(3):1407-11. | |||||
| REF 21 | Adenosine mimetics as inhibitors of NAD+-dependent histone deacetylases, from kinase to sirtuin inhibition. J Med Chem. 2006 Dec 14;49(25):7307-16. | |||||
| REF 22 | Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007 Nov 29;450(7170):712-6. | |||||
| REF 23 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Target id: 2707). | |||||
| REF 24 | Structural basis for allosteric, substrate-dependent stimulation of SIRT1 activity by resveratrol. Genes Dev. 2015 Jun 15;29(12):1316-25. | |||||
| REF 25 | Crystallographic structure of a small molecule SIRT1 activator-enzyme complex. Nat Commun. 2015 Jul 2;6:7645. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

