Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T05114
(Former ID: TTDR00197)
|
|||||
| Target Name |
Chymase (CYM)
|
|||||
| Synonyms |
Mast cell protease I; CYH; Alpha-chymase
Click to Show/Hide
|
|||||
| Gene Name |
CMA1
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 6 Target-related Diseases | + | ||||
| 1 | Atopic eczema [ICD-11: EA80] | |||||
| 2 | Chronic obstructive pulmonary disease [ICD-11: CA22] | |||||
| 3 | Gram-positive bacterial infection [ICD-11: 1B74-1F40] | |||||
| 4 | Left ventricular failure [ICD-11: BD11] | |||||
| 5 | Myocardial infarction [ICD-11: BA41-BA43] | |||||
| 6 | Heart failure [ICD-11: BD10-BD1Z] | |||||
| Function |
Major secreted protease of mast cells with suspected roles in vasoactive peptide generation, extracellular matrix degradation, and regulation of gland secretion.
Click to Show/Hide
|
|||||
| BioChemical Class |
Peptidase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 3.4.21.39
|
|||||
| Sequence |
MLLLPLPLLLFLLCSRAEAGEIIGGTECKPHSRPYMAYLEIVTSNGPSKFCGGFLIRRNF
VLTAAHCAGRSITVTLGAHNITEEEDTWQKLEVIKQFRHPKYNTSTLHHDIMLLKLKEKA SLTLAVGTLPFPSQFNFVPPGRMCRVAGWGRTGVLKPGSDTLQEVKLRLMDPQACSHFRD FDHNLQLCVGNPRKTKSAFKGDSGGPLLCAGVAQGIVSYGRSDAKPPAVFTRISHYRPWI NQILQAN Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| HIT2.0 ID | T57MP0 | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 5 Clinical Trial Drugs | + | ||||
| 1 | ASB17061 | Drug Info | Phase 2 | Atopic dermatitis | [2] | |
| 2 | BAY 11-42524 | Drug Info | Phase 2 | Left ventricular dysfunction | [3] | |
| 3 | BAY 1142524 | Drug Info | Phase 2 | Myocardial infarction | [4] | |
| 4 | JNJ-10311795 | Drug Info | Phase 2 | Chronic obstructive pulmonary disease | [5] | |
| 5 | SUN13834 | Drug Info | Phase 2 | Gram-positive bacterial infection | [6] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Modulator | [+] 2 Modulator drugs | + | ||||
| 1 | ASB17061 | Drug Info | [7] | |||
| 2 | SUN13834 | Drug Info | [10] | |||
| Inhibitor | [+] 13 Inhibitor drugs | + | ||||
| 1 | BAY 11-42524 | Drug Info | [1] | |||
| 2 | BAY 1142524 | Drug Info | [8] | |||
| 3 | JNJ-10311795 | Drug Info | [9] | |||
| 4 | 2-(N-Morpholino)-Ethanesulfonic Acid | Drug Info | [11] | |||
| 5 | BCEAB | Drug Info | [12] | |||
| 6 | Benzylsulfinic Acid | Drug Info | [13] | |||
| 7 | CHYMOSTATIN | Drug Info | [14] | |||
| 8 | KM-01221 | Drug Info | [15] | |||
| 9 | NK3201 | Drug Info | [16] | |||
| 10 | Phenylalanylmethane | Drug Info | [11] | |||
| 11 | Rac-2q | Drug Info | [14] | |||
| 12 | SUN-C8257 | Drug Info | [17] | |||
| 13 | Y-40613 | Drug Info | [18] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: (3s)-3-{3-[(6-Bromo-2-Oxo-2,3-Dihydro-1h-Indol-4-Yl)methyl]-2-Oxo-2,3-Dihydro-1h-Benzimidazol-1-Yl}hexanoic Acid | Ligand Info | |||||
| Structure Description | Crystal Structure of Human Chymase in Complex with Fragment Linked Benzimidazolone Inhibitor: (3S)-3-{3-[(6-bromo-2-oxo-2,3-dihydro-1H-indol-4-yl)methyl]-2-oxo-2,3-dihydro-1H-benzimidazol-1-yl}hexanoic acid | PDB:4K69 | ||||
| Method | X-ray diffraction | Resolution | 1.50 Å | Mutation | No | [19] |
| PDB Sequence |
IIGGTECKPH
25 SRPYMAYLEI35 VTSNGPSKFC42 GGFLIRRNFV52 LTAAHCAGRS63 ITVTLGAHNI 73 TEEEDTWQKL83 EVIKQFRHPK93 YNTSTLHHDI103 MLLKLKEKAS113 LTLAVGTLGR 134 MCRVAGWGRT144 GVLKPGSDTL155 QEVKLRLMDP165 QACSHFRDFD177 HNLQLCVGNP 185A RKTKSAFKGD194 SGGPLLCAGA208 AQGIVSYGRS218 DAKPPAVFTR230 ISHYQPWINQ 240 ILQAN
|
|||||
|
|
ILE35
4.188
LYS40
4.086
PHE41
4.183
CYS42
3.565
HIS57
3.231
CYS58
4.296
LEU99
3.992
SER189
4.343
ALA190
3.189
PHE191
3.690
LYS192
2.683
|
|||||
| Ligand Name: 6-Bromooxindole | Ligand Info | |||||
| Structure Description | Crystal Structure of Human Chymase in Complex with Fragment 6-bromo-1,3-dihydro-2H-indol-2-one | PDB:4K60 | ||||
| Method | X-ray diffraction | Resolution | 1.50 Å | Mutation | No | [19] |
| PDB Sequence |
IIGGTECKPH
25 SRPYMAYLEI35 VTNGPSKFCG43 GFLIRRNFVL53 TAAHCAGRSI64 TVTLGAHNIT 74 EEEDTWQKLE84 VIKQFRHPKY94 NTSTLHHDIM104 LLKLKEKASL114 TLAVGTLPFV 130 PPGRMCRVAG140 WGRTGVLKPG151 SDTLQEVKLR161 LMDPQACSHF173 RDFDHNLQLC 182 VGNPRKTKSA190 FKGDSGGPLL200 CAGAAQGIVS214 YGRSDAKPPA226 VFTRISHYQP 236 WINQILQAN
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Renin-angiotensin system | hsa04614 | Affiliated Target |
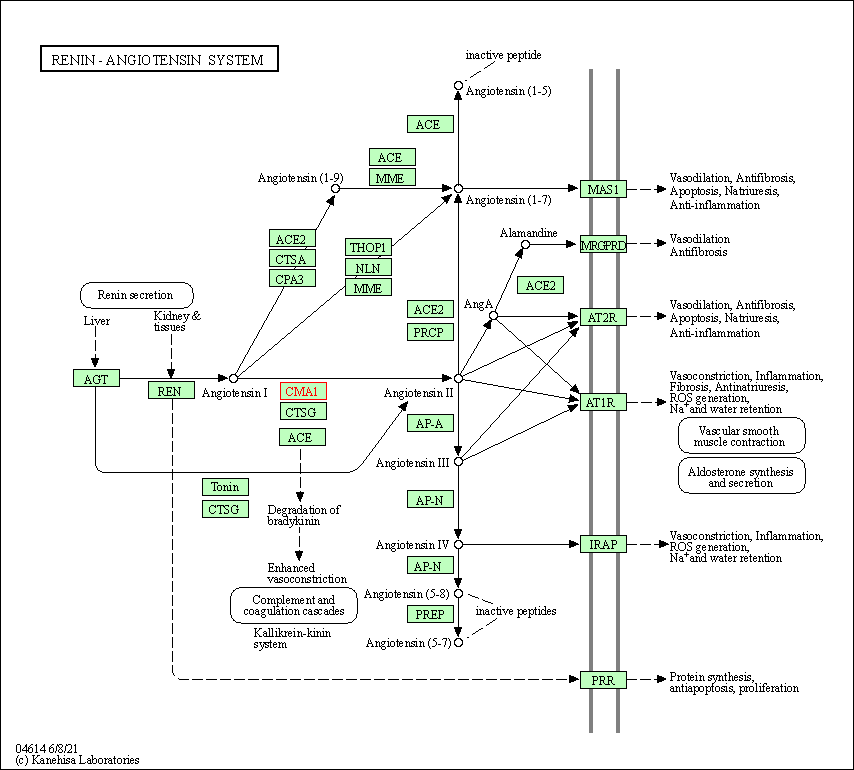
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Degree | 2 | Degree centrality | 2.15E-04 | Betweenness centrality | 2.50E-06 |
|---|---|---|---|---|---|
| Closeness centrality | 1.89E-01 | Radiality | 1.32E+01 | Clustering coefficient | 0.00E+00 |
| Neighborhood connectivity | 1.30E+01 | Topological coefficient | 5.00E-01 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 1 KEGG Pathways | + | ||||
| 1 | Renin-angiotensin system | |||||
| Reactome | [+] 2 Reactome Pathways | + | ||||
| 1 | Activation of Matrix Metalloproteinases | |||||
| 2 | Metabolism of Angiotensinogen to Angiotensins | |||||
| WikiPathways | [+] 2 WikiPathways | + | ||||
| 1 | ACE Inhibitor Pathway | |||||
| 2 | Metabolism of Angiotensinogen to Angiotensins | |||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | ClinicalTrials.gov (NCT02452515) A Single-blind Pilot Study to Investigate Safety and Tolerability of the Chymase Inhibitor BAY1142524 in Clinically Stable Patients With Left-ventricular Dysfunction. | |||||
| REF 2 | ClinicalTrials.gov (NCT01756898) A Study to Evaluate the Efficacy, Safety and Pharmacokinetics of ASB17061 Capsules in Adult Subjects With Atopic Dermatitis. U.S. National Institutes of Health. | |||||
| REF 3 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 4 | ClinicalTrials.gov (NCT02976467) A Double-blind Study to Investigate Efficacy, Safety and Tolerability of BAY 1142524 in Patients After Acute Myocardial Infarction With Left-ventricular Dysfunction (CHIARA MIA 2). U.S. National Institutes of Health. | |||||
| REF 5 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 6563). | |||||
| REF 6 | Emerging drugs for bacterial urinary tract infections. Expert Opin Emerg Drugs. 2005 May;10(2):275-98. | |||||
| REF 7 | Immunology of atopic eczema: overcoming the Th1/Th2 paradigm. Allergy. 2013 Aug;68(8):974-82. | |||||
| REF 8 | Pharmacokinetics, Safety, and Tolerability of the Novel Chymase Inhibitor BAY 1142524 in Healthy Male Volunteers. Clin Pharmacol Drug Dev. 2019 May;8(4):467-479. | |||||
| REF 9 | Discovery of potent, selective, orally active, nonpeptide inhibitors of human mast cell chymase. J Med Chem. 2007 Apr 19;50(8):1727-30. | |||||
| REF 10 | Oral chymase inhibitor SUN13834 ameliorates skin inflammation as well as pruritus in mouse model for atopic dermatitis.Eur J Pharmacol.2008 Dec 28;601(1-3):186-91. | |||||
| REF 11 | How many drug targets are there Nat Rev Drug Discov. 2006 Dec;5(12):993-6. | |||||
| REF 12 | Development of a chymase inhibitor: pharmacological characterization of a chymase inhibitor in inflamed tissue remodeling and fibrosis. Jpn J Pharmacol. 2002 Nov;90(3):206-9. | |||||
| REF 13 | The Protein Data Bank. Nucleic Acids Res. 2000 Jan 1;28(1):235-42. | |||||
| REF 14 | Identification of 6-substituted 4-arylsulfonyl-1,4-diazepane-2,5-diones as a novel scaffold for human chymase inhibitors. Bioorg Med Chem Lett. 2007 Jun 15;17(12):3431-4. | |||||
| REF 15 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Target id: 2340). | |||||
| REF 16 | Impact of chymase inhibitor on cardiac function and survival after myocardial infarction. Cardiovasc Res. 2003 Nov 1;60(2):413-20. | |||||
| REF 17 | Chymase inhibitor ameliorates eosinophilia in mice infected with Nippostrongylus brasiliensis. Int Arch Allergy Immunol. 2002 Jul;128(3):235-9. | |||||
| REF 18 | Therapeutic potential of a specific chymase inhibitor in atopic dermatitis. Jpn J Pharmacol. 2002 Nov;90(3):214-7. | |||||
| REF 19 | Discovery of potent, selective chymase inhibitors via fragment linking strategies. J Med Chem. 2013 Jun 13;56(11):4465-81. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

