| Drug General Information |
| Drug ID |
D0UU1I
|
| Former ID |
DNC002423
|
| Drug Name |
Heme
|
| Drug Type |
Small molecular drug
|
| Indication |
Discovery agent
|
Investigative |
[1]
|
|---|
| Structure |
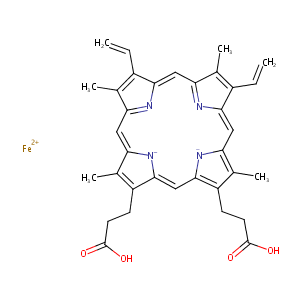
|
Download
2D MOL
3D MOL
|
| Formula |
C34H32FeN4O4
|
| InChI |
InChI=1S/C34H34N4O4.Fe/c1-7-21-17(3)25-13-26-19(5)23(9-11-33(39)40)31(37-26)16-32-24(10-12-34(41)42)20(6)28(38-32)15-30-22(8-2)18(4)27(36-30)14-29(21)35-25;/h7-8,13-16H,1-2,9-12H2,3-6H3,(H4,35,36,37,38,39,40,41,42);/q;+2/p-2
|
| InChIKey |
KABFMIBPWCXCRK-UHFFFAOYSA-L
|
| PubChem Compound ID |
|
| PubChem Substance ID |
598147, 11567731, 11567732, 50074840, 53790568, 57310353, 57322552, 76721894, 104298682, 104307920, 104631336, 104631339, 134989497, 160966170, 163113594, 178101166, 198981484
|
| Target and Pathway |
| Target(s) |
Flavohemoglobin |
Target Info |
Inhibitor |
[2]
|
|---|
| Heme oxygenase |
Target Info |
Inhibitor |
[2]
|
| Prostaglandin G/H synthase 1 |
Target Info |
Inhibitor |
[2]
|
| Fumarate reductase flavoprotein subunit |
Target Info |
Inhibitor |
[2]
|
| Prostaglandin G/H synthase 2 |
Target Info |
Inhibitor |
[2]
|
| Cytochrome p450 121 |
Target Info |
Inhibitor |
[2]
|
| Cystathionine beta-synthase |
Target Info |
Inhibitor |
[2]
|
| Arachidonate 5-lipoxygenase |
Target Info |
Inhibitor |
[2]
|
| Nitric-oxide synthase, endothelial |
Target Info |
Inhibitor |
[2]
|
| Hemoglobin |
Target Info |
Inhibitor |
[2]
|
| Nitric-oxide synthase, brain |
Target Info |
Inhibitor |
[2]
|
| Cytochrome P450 3A4 |
Target Info |
Inhibitor |
[2]
|
| Nitric oxide synthase, inducible |
Target Info |
Inhibitor |
[2]
|
|
BioCyc Pathway
|
Heme degradationPWY66-374:C20 prostanoid biosynthesisPWY66-393:Aspirin-triggered lipoxin biosynthesis
|
|
Aspirin triggered resolvin D biosynthesis
|
|
C20 prostanoid biosynthesis
|
|
Aspirin triggered resolvin E biosynthesisPWY66-426:Hydrogen sulfide biosynthesis (trans-sulfuration)
|
|
Cysteine biosynthesis/homocysteine degradation (trans-sulfuration)
|
|
Superpathway of methionine degradation
|
|
Cysteine biosynthesisPWY66-393:Aspirin-triggered lipoxin biosynthesis
|
|
Resolvin D biosynthesis
|
|
Leukotriene biosynthesis
|
|
Lipoxin biosynthesis
|
|
Aspirin triggered resolvin E biosynthesisPWY-4983:Citrulline-nitric oxide cyclePWY-4983:Citrulline-nitric oxide cyclePWY-4983:Citrulline-nitric oxide cycle
|
|
KEGG Pathway
|
Porphyrin and chlorophyll metabolism
|
|
HIF-1 signaling pathway
|
|
Mineral absorption
|
|
MicroRNAs in cancerhsa00590:Arachidonic acid metabolism
|
|
Metabolic pathways
|
|
Platelet activation
|
|
Serotonergic synapsehsa00590:Arachidonic acid metabolism
|
|
NF-kappa B signaling pathway
|
|
VEGF signaling pathway
|
|
TNF signaling pathway
|
|
Retrograde endocannabinoid signaling
|
|
Serotonergic synapse
|
|
Ovarian steroidogenesis
|
|
Oxytocin signaling pathway
|
|
Regulation of lipolysis in adipocytes
|
|
Leishmaniasis
|
|
Pathways in cancer
|
|
Chemical carcinogenesis
|
|
MicroRNAs in cancer
|
|
Small cell lung cancerhsa00260:Glycine, serine and threonine metabolism
|
|
Cysteine and methionine metabolism
|
|
Biosynthesis of antibiotics
|
|
Biosynthesis of amino acidshsa00590:Arachidonic acid metabolism
|
|
Toxoplasmosishsa00330:Arginine and proline metabolism
|
|
Calcium signaling pathway
|
|
cGMP-PKG signaling pathway
|
|
Sphingolipid signaling pathway
|
|
PI3K-Akt signaling pathway
|
|
Estrogen signaling pathway
|
|
Oxytocin signaling pathwayhsa05143:African trypanosomiasis
|
|
Malariahsa00330:Arginine and proline metabolism
|
|
Phagosome
|
|
Circadian entrainment
|
|
Long-term depression
|
|
Salivary secretion
|
|
Alzheimer's disease
|
|
Amyotrophic lateral sclerosis (ALS)hsa00140:Steroid hormone biosynthesis
|
|
Linoleic acid metabolism
|
|
Retinol metabolism
|
|
Metabolism of xenobiotics by cytochrome P450
|
|
Drug metabolism - cytochrome P450
|
|
Drug metabolism - other enzymes
|
|
Bile secretion
|
|
Chemical carcinogenesishsa00220:Arginine biosynthesis
|
|
Arginine and proline metabolism
|
|
Peroxisome
|
|
Salmonella infection
|
|
Pertussis
|
|
Chagas disease (American trypanosomiasis)
|
|
Toxoplasmosis
|
|
Amoebiasis
|
|
Tuberculosis
|
|
Small cell lung cancer
|
|
NetPath Pathway
|
TGF_beta_Receptor Signaling PathwayNetPath_13:IL1 Signaling Pathway
|
|
TSH Signaling Pathway
|
|
IL4 Signaling Pathway
|
|
TGF_beta_Receptor Signaling Pathway
|
|
IL5 Signaling PathwayNetPath_7:TGF_beta_Receptor Signaling PathwayNetPath_16:IL4 Signaling PathwayNetPath_8:Wnt Signaling PathwayNetPath_4:EGFR1 Signaling PathwayNetPath_13:IL1 Signaling Pathway
|
|
IL2 Signaling Pathway
|
|
PANTHER Pathway
|
Inflammation mediated by chemokine and cytokine signaling pathwayP00019:Endothelin signaling pathway
|
|
Inflammation mediated by chemokine and cytokine signaling pathway
|
|
Toll receptor signaling pathway
|
|
CCKR signaling map STP02737:Cysteine biosynthesisP00005:Angiogenesis
|
|
Endothelin signaling pathway
|
|
Interleukin signaling pathway
|
|
PI3 kinase pathway
|
|
VEGF signaling pathwayP06959:CCKR signaling map ST
|
|
Pathway Interaction Database
|
Validated transcriptional targets of AP1 family members Fra1 and Fra2
|
|
HIF-1-alpha transcription factor networknfat_tfpathway:Calcineurin-regulated NFAT-dependent transcription in lymphocytes
|
|
S1P1 pathway
|
|
C-MYB transcription factor network
|
|
Signaling mediated by p38-alpha and p38-beta
|
|
Calcium signaling in the CD4+ TCR pathwayer_nongenomic_pathway:Plasma membrane estrogen receptor signaling
|
|
Angiopoietin receptor Tie2-mediated signaling
|
|
Thromboxane A2 receptor signaling
|
|
SHP2 signaling
|
|
VEGFR1 specific signals
|
|
Signaling events mediated by VEGFR1 and VEGFR2
|
|
PAR1-mediated thrombin signaling eventsil12_2pathway:IL12-mediated signaling events
|
|
Alpha9 beta1 integrin signaling events
|
|
ATF-2 transcription factor network
|
|
IL23-mediated signaling events
|
|
HIF-1-alpha transcription factor network
|
|
PathWhiz Pathway
|
Porphyrin MetabolismPW000044:Arachidonic Acid MetabolismPW000044:Arachidonic Acid MetabolismPW000007:Selenoamino Acid Metabolism
|
|
Homocysteine Degradation
|
|
Methionine Metabolism
|
|
Glycine and Serine MetabolismPW000044:Arachidonic Acid MetabolismPW000010:Arginine and Proline MetabolismPW000015:Caffeine Metabolism
|
|
Retinol Metabolism
|
|
Reactome
|
Iron uptake and transportR-HSA-1222556:ROS production in response to bacteria
|
|
Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation
|
|
eNOS activation
|
|
Nitric oxide stimulates guanylate cyclase
|
|
VEGFR2 mediated vascular permeabilityR-HSA-1237044:Erythrocytes take up carbon dioxide and release oxygen
|
|
Erythrocytes take up oxygen and release carbon dioxide
|
|
Scavenging of heme from plasmaR-HSA-1222556:ROS production in response to bacteria
|
|
Nitric oxide stimulates guanylate cyclaseR-HSA-211981:Xenobiotics
|
|
Aflatoxin activation and detoxificationR-HSA-1222556:ROS production in response to bacteria
|
|
WikiPathways
|
Oxidative Stress
|
|
Transcriptional activation by NRF2
|
|
NRF2 pathway
|
|
Nuclear Receptors Meta-Pathway
|
|
miR-targeted genes in squamous cell - TarBase
|
|
miR-targeted genes in muscle cell - TarBase
|
|
miR-targeted genes in lymphocytes - TarBase
|
|
miR-targeted genes in leukocytes - TarBase
|
|
miR-targeted genes in epithelium - TarBaseWP98:Prostaglandin Synthesis and Regulation
|
|
Arachidonic acid metabolism
|
|
Phase 1 - Functionalization of compounds
|
|
Eicosanoid Synthesis
|
|
Selenium Micronutrient NetworkWP98:Prostaglandin Synthesis and Regulation
|
|
Aryl Hydrocarbon Receptor
|
|
Quercetin and Nf-kB/ AP-1 Induced Cell Apoptosis
|
|
Spinal Cord Injury
|
|
Integrated Pancreatic Cancer Pathway
|
|
Selenium Micronutrient NetworkWP2877:Vitamin D Receptor Pathway
|
|
Metabolism of amino acids and derivatives
|
|
Trans-sulfuration and one carbon metabolism
|
|
Trans-sulfuration pathway
|
|
Folate Metabolism
|
|
Folate-Alcohol and Cancer Pathway
|
|
Vitamin B12 Metabolism
|
|
Selenium Micronutrient NetworkWP554:ACE Inhibitor Pathway
|
|
EGF/EGFR Signaling Pathway
|
|
Myometrial Relaxation and Contraction Pathways
|
|
JAK/STAT
|
|
Corticotropin-releasing hormone
|
|
AGE/RAGE pathway
|
|
Endothelin Pathways
|
|
Leptin signaling pathway
|
|
Effects of Nitric Oxide
|
|
Metabolism of nitric oxide
|
|
AngiogenesisWP2784:Binding and Uptake of Ligands by Scavenger Receptors
|
|
Uptake of Carbon Dioxide and Release of Oxygen by Erythrocytes
|
|
Uptake of Oxygen and Release of Carbon Dioxide by Erythrocytes
|
|
Factors involved in megakaryocyte development and platelet production
|
|
Selenium Micronutrient NetworkWP727:Monoamine Transport
|
|
Amyotrophic lateral sclerosis (ALS)
|
|
Alzheimers Disease
|
|
Serotonin Transporter ActivityWP702:Metapathway biotransformation
|
|
Aflatoxin B1 metabolism
|
|
Estrogen metabolism
|
|
Benzo(a)pyrene metabolism
|
|
Tamoxifen metabolism
|
|
Tryptophan metabolism
|
|
Oxidation by Cytochrome P450
|
|
Nuclear Receptors in Lipid Metabolism and Toxicity
|
|
Farnesoid X Receptor Pathway
|
|
Vitamin D Receptor Pathway
|
|
Felbamate Metabolism
|
|
Lidocaine metabolism
|
|
Nifedipine Activity
|
|
Colchicine Metabolic Pathway
|
|
Irinotecan Pathway
|
|
Drug Induction of Bile Acid Pathway
|
|
Fatty Acid Omega Oxidation
|
|
Codeine and Morphine MetabolismWP619:Type II interferon signaling (IFNG)
|
| References |
| REF 1 | (http://www.guidetopharmacology.org/) Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 4349). |
|---|
| REF 2 | The Protein Data Bank. Nucleic Acids Res. 2000 Jan 1;28(1):235-42. |
|---|