Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T90439
(Former ID: TTDI00151)
|
|||||
| Target Name |
Jagged-2 protein (JAG2)
|
|||||
| Synonyms |
hJ2; Protein jagged-2; Jagged2
Click to Show/Hide
|
|||||
| Gene Name |
JAG2
|
|||||
| Target Type |
Literature-reported target
|
[1] | ||||
| Function |
Involved in limb development. Putative Notch ligand involved in the mediation of Notch signaling.
Click to Show/Hide
|
|||||
| UniProt ID | ||||||
| Sequence |
MRAQGRGRLPRRLLLLLALWVQAARPMGYFELQLSALRNVNGELLSGACCDGDGRTTRAG
GCGHDECDTYVRVCLKEYQAKVTPTGPCSYGHGATPVLGGNSFYLPPAGAAGDRARARAR AGGDQDPGLVVIPFQFAWPRSFTLIVEAWDWDNDTTPNEELLIERVSHAGMINPEDRWKS LHFSGHVAHLELQIRVRCDENYYSATCNKFCRPRNDFFGHYTCDQYGNKACMDGWMGKEC KEAVCKQGCNLLHGGCTVPGECRCSYGWQGRFCDECVPYPGCVHGSCVEPWQCNCETNWG GLLCDKDLNYCGSHHPCTNGGTCINAEPDQYRCTCPDGYSGRNCEKAEHACTSNPCANGG SCHEVPSGFECHCPSGWSGPTCALDIDECASNPCAAGGTCVDQVDGFECICPEQWVGATC QLDANECEGKPCLNAFSCKNLIGGYYCDCIPGWKGINCHINVNDCRGQCQHGGTCKDLVN GYQCVCPRGFGGRHCELERDECASSPCHSGGLCEDLADGFHCHCPQGFSGPLCEVDVDLC EPSPCRNGARCYNLEGDYYCACPDDFGGKNCSVPREPCPGGACRVIDGCGSDAGPGMPGT AASGVCGPHGRCVSQPGGNFSCICDSGFTGTYCHENIDDCLGQPCRNGGTCIDEVDAFRC FCPSGWEGELCDTNPNDCLPDPCHSRGRCYDLVNDFYCACDDGWKGKTCHSREFQCDAYT CSNGGTCYDSGDTFRCACPPGWKGSTCAVAKNSSCLPNPCVNGGTCVGSGASFSCICRDG WEGRTCTHNTNDCNPLPCYNGGICVDGVNWFRCECAPGFAGPDCRINIDECQSSPCAYGA TCVDEINGYRCSCPPGRAGPRCQEVIGFGRSCWSRGTPFPHGSSWVEDCNSCRCLDGRRD CSKVWCGWKPCLLAGQPEALSAQCPLGQRCLEKAPGQCLRPPCEAWGECGAEEPPSTPCL PRSGHLDNNCARLTLHFNRDHVPQGTTVGAICSGIRSLPATRAVARDRLLVLLCDRASSG ASAVEVAVSFSPARDLPDSSLIQGAAHAIVAAITQRGNSSLLLAVTEVKVETVVTGGSST GLLVPVLCGAFSVLWLACVVLCVWWTRKRRKERERSRLPREESANNQWAPLNPIRNPIER PGGHKDVLYQCKNFTPPPRRADEALPGPAGHAAVREDEEDEDLGRGEEDSLEAEKFLSHK FTKDPGRSPGRPAHWASGPKVDNRAVRSINEARYAGKE Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: alpha-L-Fucose | Ligand Info | |||||
| Structure Description | Human Jagged2 C2-EGF3 | PDB:5MW7 | ||||
| Method | X-ray diffraction | Resolution | 2.80 Å | Mutation | No | [2] |
| PDB Sequence |
MGYFELQLSA
36 LRNVNGELLS46 GACCDCDECD68 TYVRVCLKEY78 QAKVTPTGPC88 SYGHGATPVL 98 GGNSFYLPPA108 GAAGDRARAR118 ARAGGDQDPG128 LVVIPFQFAW138 PRSFTLIVEA 148 WDWDNNEELL162 IERVSHAGMI172 NPEDRWKSLH182 FSGHVAHLEL192 QIRVRCDENY 202 YSATCNKFCR212 PRNDFFGHYT222 CDQYGNKACM232 DGWMGKECKE242 AVCKQGCNLL 252 HGGCTVPGEC262 RCSYGWQGRF272 CDECVPYPGC282 VHGSCVEPWQ292 CNCETNWGGL 302 LCDKDLNYCG312 SHHPCTNGGT322 CINAEPDQYR332 CT
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Pathway Affiliation
Biological Network Descriptors
|
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Notch signaling pathway | hsa04330 | Affiliated Target |
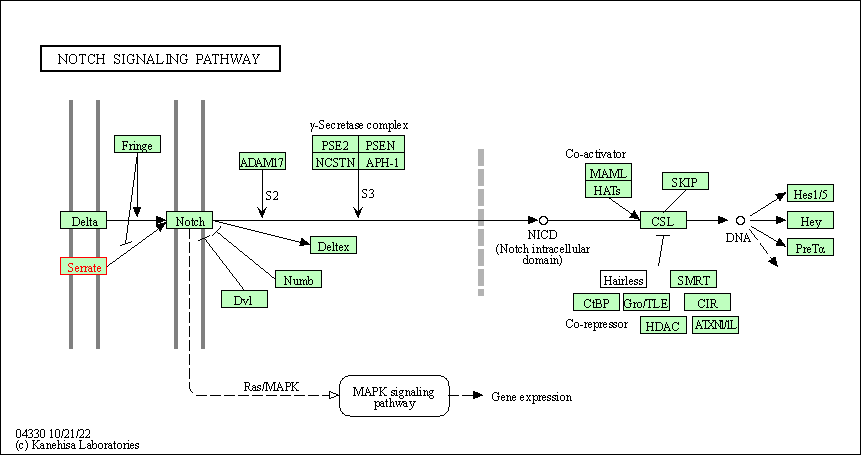
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Th1 and Th2 cell differentiation | hsa04658 | Affiliated Target |
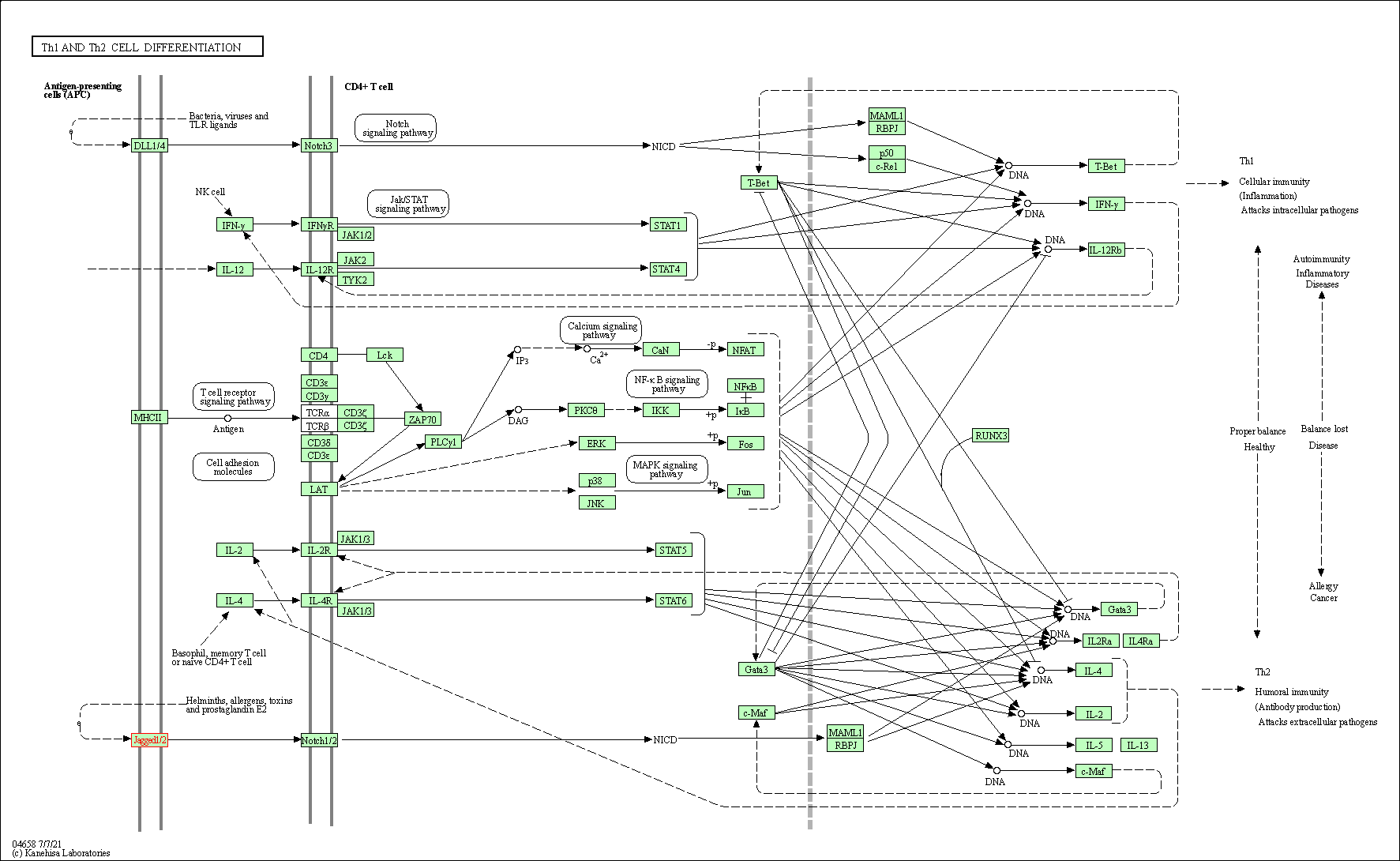
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Degree | 8 | Degree centrality | 8.59E-04 | Betweenness centrality | 2.51E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 2.04E-01 | Radiality | 1.36E+01 | Clustering coefficient | 2.86E-01 |
| Neighborhood connectivity | 1.71E+01 | Topological coefficient | 2.28E-01 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | A microRNA-1280/JAG2 network comprises a novel biological target in high-risk medulloblastoma. Oncotarget. 2015 Feb 20;6(5):2709-24. | |||||
| REF 2 | Structural and functional dissection of the interplay between lipid and Notch binding by human Notch ligands. EMBO J. 2017 Aug 1;36(15):2204-2215. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

