Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T82795
(Former ID: TTDR00713)
|
|||||
| Target Name |
Beta-catenin (CTNNB1)
|
|||||
| Synonyms |
Wnt/beta-catenin signaling pathway; PRO2286; OK/SW-cl.35; Catenin beta-1; CTNNB
Click to Show/Hide
|
|||||
| Gene Name |
CTNNB1
|
|||||
| Target Type |
Successful target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||||
| Function |
In the absence of Wnt, forms a complex with AXIN1, AXIN2, APC, CSNK1A1 and GSK3B that promotes phosphorylation on N-terminal Ser and Thr residues and ubiquitination of CTNNB1 via BTRC and its subsequent degradation by the proteasome. In the presence of Wnt ligand, CTNNB1 is not ubiquitinated and accumulates in the nucleus, where it acts as a coactivator for transcription factors of the TCF/LEF family, leading to activate Wnt responsive genes. Involved in the regulation of cell adhesion, as component of an E-cadherin:catenin adhesion complex. Acts as a negative regulator of centrosome cohesion. Involved in the CDK2/PTPN6/CTNNB1/CEACAM1 pathway of insulin internalization. Blocks anoikis of malignant kidney and intestinal epithelial cells and promotes their anchorage-independent growth by down-regulating DAPK2. Disrupts PML function and PML-NB formation by inhibiting RANBP2-mediated sumoylation of PML. Promotes neurogenesis by maintaining sympathetic neuroblasts within the cell cycle. Key downstream component of the canonical Wnt signaling pathway.
Click to Show/Hide
|
|||||
| BioChemical Class |
Beta-catenin
|
|||||
| UniProt ID | ||||||
| Sequence |
MATQADLMELDMAMEPDRKAAVSHWQQQSYLDSGIHSGATTTAPSLSGKGNPEEEDVDTS
QVLYEWEQGFSQSFTQEQVADIDGQYAMTRAQRVRAAMFPETLDEGMQIPSTQFDAAHPT NVQRLAEPSQMLKHAVVNLINYQDDAELATRAIPELTKLLNDEDQVVVNKAAVMVHQLSK KEASRHAIMRSPQMVSAIVRTMQNTNDVETARCTAGTLHNLSHHREGLLAIFKSGGIPAL VKMLGSPVDSVLFYAITTLHNLLLHQEGAKMAVRLAGGLQKMVALLNKTNVKFLAITTDC LQILAYGNQESKLIILASGGPQALVNIMRTYTYEKLLWTTSRVLKVLSVCSSNKPAIVEA GGMQALGLHLTDPSQRLVQNCLWTLRNLSDAATKQEGMEGLLGTLVQLLGSDDINVVTCA AGILSNLTCNNYKNKMMVCQVGGIEALVRTVLRAGDREDITEPAICALRHLTSRHQEAEM AQNAVRLHYGLPVVVKLLHPPSHWPLIKATVGLIRNLALCPANHAPLREQGAIPRLVQLL VRAHQDTQRRTSMGGTQQQFVEGVRMEEIVEGCTGALHILARDVHNRIVIRGLNTIPLFV QLLYSPIENIQRVAAGVLCELAQDKEAAEAIEAEGATAPLTELLHSRNEGVATYAAAVLF RMSEDKPQDYKKRLSVELTSSLFRTEPMAWNETADLGLDIGAQGEPLGYRQDDPSYRSFH SGGYGQDALGMDPMMEHEMGGHHPGADYPVDGLPDLGHAQDLMDGLPPGDSNQLAWFDTD L Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| ADReCS ID | BADD_A02497 ; BADD_A05554 | |||||
| HIT2.0 ID | T92BZ2 | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Approved Drug(s) | [+] 1 Approved Drugs | + | ||||
| 1 | Recombinant human endostatin | Drug Info | Approved | Solid tumour/cancer | [2] | |
| Clinical Trial Drug(s) | [+] 3 Clinical Trial Drugs | + | ||||
| 1 | C 82 | Drug Info | Phase 1/2 | Scleroderma | [3] | |
| 2 | CEQ-508 | Drug Info | Phase 1/2 | Familial adenomatous polyposis | [4] | |
| 3 | Tegavivint | Drug Info | Phase 1 | Desmoid tumour | [5] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Modulator | [+] 1 Modulator drugs | + | ||||
| 1 | Recombinant human endostatin | Drug Info | [1], [2] | |||
| Inhibitor | [+] 2 Inhibitor drugs | + | ||||
| 1 | C 82 | Drug Info | [3] | |||
| 2 | Tegavivint | Drug Info | [7] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: L-serine-O-phosphate | Ligand Info | |||||
| Structure Description | Monophosphorylated pSer33 b-Catenin peptide, b-TrCP/Skp1, NRX-2776 ternary complex | PDB:6M90 | ||||
| Method | X-ray diffraction | Resolution | 2.05 Å | Mutation | No | [8] |
| PDB Sequence |
LDGIHSGAT
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Ligand Name: 3-[(2~{R})-4-methyl-5-oxidanylidene-2,3-dihydro-1,4-benzoxazepin-2-yl]benzenecarbonitrile | Ligand Info | |||||
| Structure Description | Beta-Catenin in complex with compound 6 | PDB:7AFW | ||||
| Method | X-ray diffraction | Resolution | 1.81 Å | Mutation | No | [9] |
| PDB Sequence |
DDAELATRAI
153 PELTKLLNDE163 DQVVVNKAAV173 MVHQLSKKEA183 SRHAIMRSPQ193 MVSAIVRTMQ 203 NTNDVETARC213 TAGTLHNLSH223 HREGLLAIFK233 SGGIPALVKM243 LGSPVDSVLF 253 YAITTLHNLL263 LHQEGAKMAV273 RLAGGLQKMV283 ALLNKTNVKF293 LAITTDCLQI 303 L
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Rap1 signaling pathway | hsa04015 | Affiliated Target |
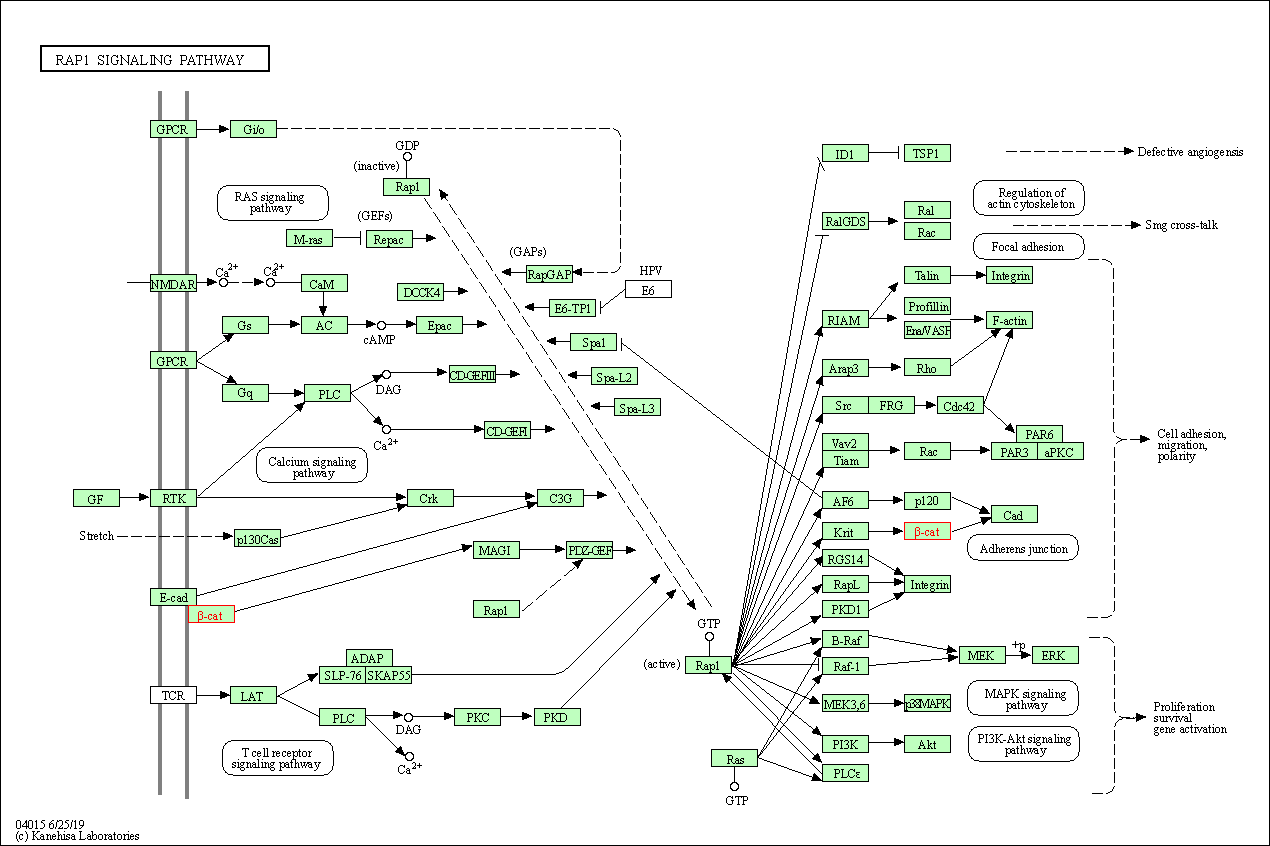
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Wnt signaling pathway | hsa04310 | Affiliated Target |
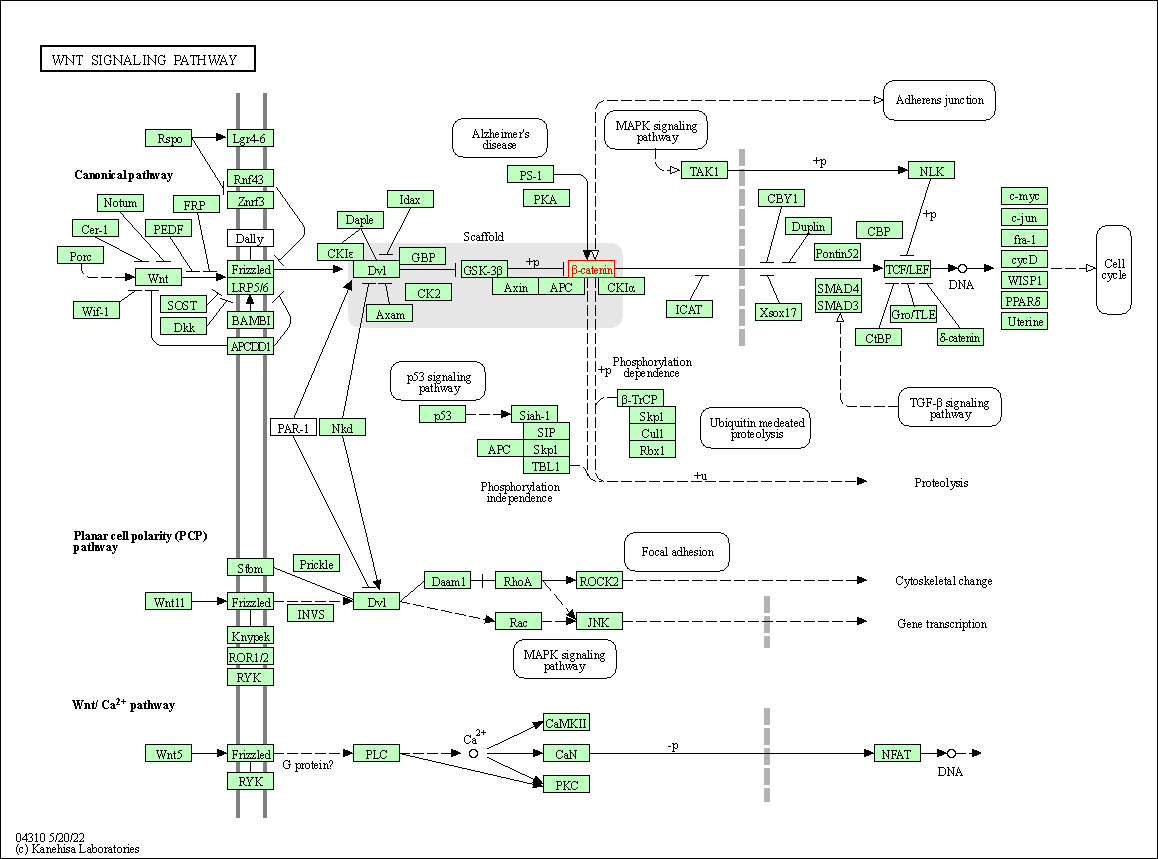
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Hippo signaling pathway | hsa04390 | Affiliated Target |
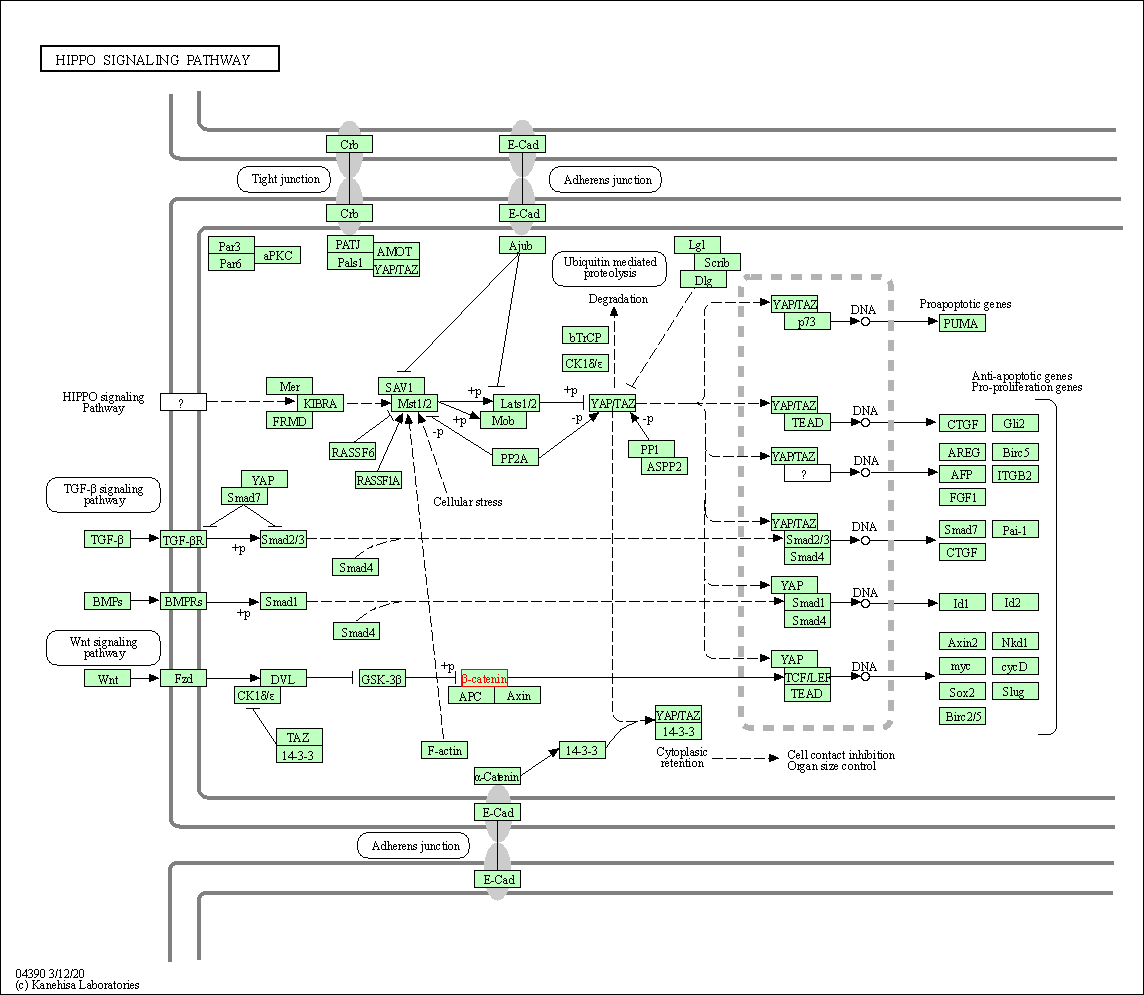
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Focal adhesion | hsa04510 | Affiliated Target |
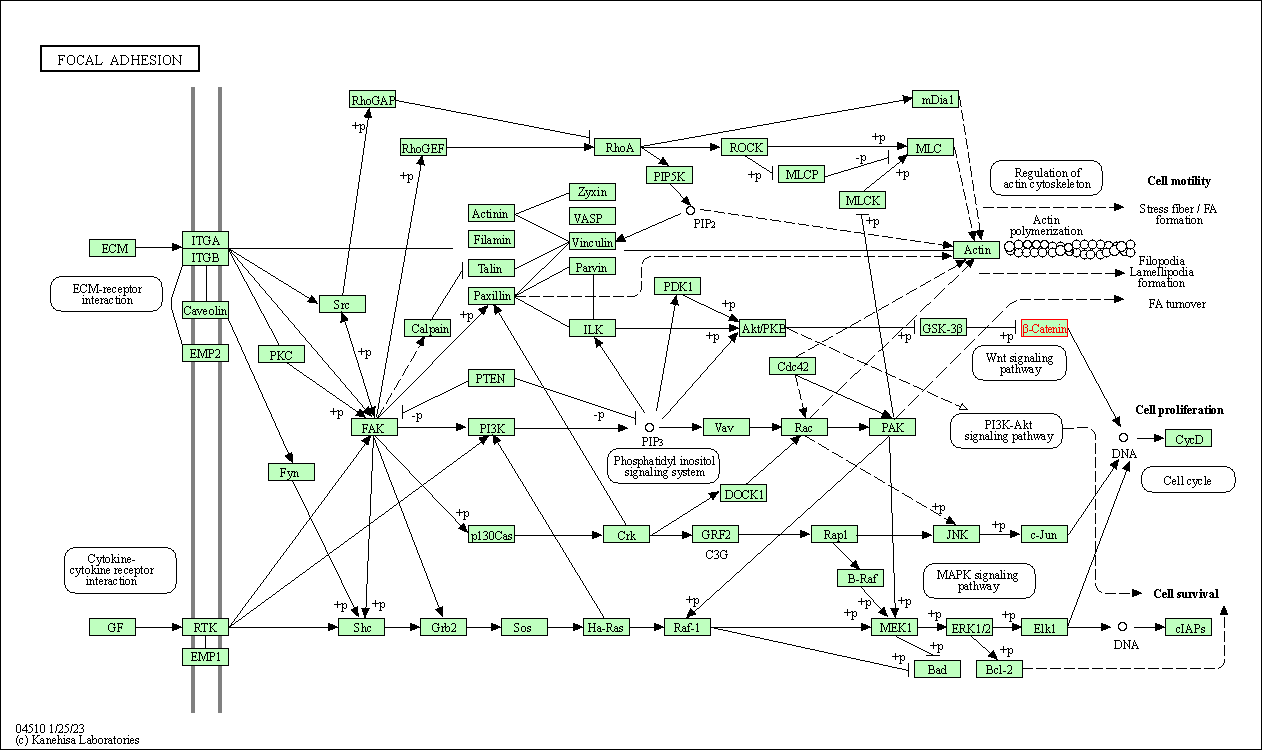
|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
| Adherens junction | hsa04520 | Affiliated Target |
|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
| Signaling pathways regulating pluripotency of stem cells | hsa04550 | Affiliated Target |
|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
| Leukocyte transendothelial migration | hsa04670 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Melanogenesis | hsa04916 | Affiliated Target |
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Thyroid hormone signaling pathway | hsa04919 | Affiliated Target |
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 209 | Degree centrality | 2.25E-02 | Betweenness centrality | 5.62E-02 |
|---|---|---|---|---|---|
| Closeness centrality | 2.93E-01 | Radiality | 1.49E+01 | Clustering coefficient | 3.70E-02 |
| Neighborhood connectivity | 2.96E+01 | Topological coefficient | 1.38E-02 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Endostar, a modified recombinant human endostatin, suppresses angiogenesis through inhibition of Wnt/beta-catenin signaling pathway. PLoS One. 2014 Sep 18;9(9):e107463. | |||||
| REF 2 | Endostar, a modified recombinant human endostatin, suppresses angiogenesis through inhibition of Wnt/beta-catenin signaling pathway. PLoS One. 2014 Sep 18;9(9):e107463. | |||||
| REF 3 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 4 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 5 | ClinicalTrials.gov (NCT03459469) Phase 1 Trial of BC2059 (Tegavivint) in Patients With Unresectable Desmoid Tumor. U.S.National Institutes of Health. | |||||
| REF 6 | Current Progress of siRNA/shRNA Therapeutics in Clinical Trials. Biotechnol J. 2011 September; 6(9): 1130-1146. | |||||
| REF 7 | Tegavivint and the beta-Catenin/ALDH Axis in Chemotherapy-Resistant and Metastatic Osteosarcoma. J Natl Cancer Inst. 2019 Nov 1;111(11):1216-1227. | |||||
| REF 8 | Prospective discovery of small molecule enhancers of an E3 ligase-substrate interaction. Nat Commun. 2019 Mar 29;10(1):1402. | |||||
| REF 9 | Getting a Grip on the Undrugged: Targeting beta-Catenin with Fragment-Based Methods. ChemMedChem. 2021 May 6;16(9):1420-1424. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

