Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T78915
(Former ID: TTDI01969)
|
|||||
| Target Name |
Tryptophan 5-hydroxylase 1 (TPH1)
|
|||||
| Synonyms |
Tryptophan 5-monooxygenase 1; TRPH; TPRH
Click to Show/Hide
|
|||||
| Gene Name |
TPH1
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 2 Target-related Diseases | + | ||||
| 1 | Psychotic disorder [ICD-11: 6A20-6A25] | |||||
| 2 | Pulmonary hypertension [ICD-11: BB01] | |||||
| Function |
Responsible for addition of the -HO group (hydroxylation) to the 5 position to form the amino acid 5-hydroxytryptophan (5-HTP), which is the initial and rate-limiting step in the synthesis of the neurotransmitter serotonin.
Click to Show/Hide
|
|||||
| BioChemical Class |
Paired donor oxygen oxidoreductase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 1.14.16.4
|
|||||
| Sequence |
MIEDNKENKDHSLERGRASLIFSLKNEVGGLIKALKIFQEKHVNLLHIESRKSKRRNSEF
EIFVDCDINREQLNDIFHLLKSHTNVLSVNLPDNFTLKEDGMETVPWFPKKISDLDHCAN RVLMYGSELDADHPGFKDNVYRKRRKYFADLAMNYKHGDPIPKVEFTEEEIKTWGTVFQE LNKLYPTHACREYLKNLPLLSKYCGYREDNIPQLEDVSNFLKERTGFSIRPVAGYLSPRD FLSGLAFRVFHCTQYVRHSSDPFYTPEPDTCHELLGHVPLLAEPSFAQFSQEIGLASLGA SEEAVQKLATCYFFTVEFGLCKQDGQLRVFGAGLLSSISELKHALSGHAKVKPFDPKITC KQECLITTFQDVYFVSESFEDAKEKMREFTKTIKRPFGVKYNPYTRSIQILKDTKSITSA MNELQHDLDVVSDALAKVSRKPSI Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| HIT2.0 ID | T39T57 | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 2 Clinical Trial Drugs | + | ||||
| 1 | AGN-2979 | Drug Info | Phase 2 | Psychotic disorder | [1] | |
| 2 | KAR5585 | Drug Info | Phase 1 | Pulmonary arterial hypertension | [2] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Modulator | [+] 1 Modulator drugs | + | ||||
| 1 | AGN-2979 | Drug Info | [1] | |||
| Inhibitor | [+] 3 Inhibitor drugs | + | ||||
| 1 | 6-fluorotryptophan | Drug Info | [3] | |||
| 2 | alpha-propyldopacetamide | Drug Info | [4] | |||
| 3 | fenclonine | Drug Info | [4] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: KAR5585 | Ligand Info | |||||
| Structure Description | Tryptophan 5-hydroxylase in complex with inhibitor (3~{S})-8-[2-azanyl-6-[(1~{R})-1-(4-chloranyl-2-phenyl-phenyl)-2,2,2-tris(fluoranyl)ethoxy]pyrimidin-4-yl]-2,8-diazaspiro[4.5]decane-3-carboxylic acid | PDB:5L01 | ||||
| Method | X-ray diffraction | Resolution | 1.90 Å | Mutation | No | [5] |
| PDB Sequence |
TVPWFPKKIS
113 DLDHCANRVL123 MYGGFKDNVY141 RKRRKYFADL151 AMNYKHGDPI161 PKVEFTEEEI 171 KTWGTVFQEL181 NKLYPTHACR191 EYLKNLPLLS201 KYCGYREDNI211 PQLEDVSNFL 221 KERTGFSIRP231 VAGYLSPRDF241 LSGLAFRVFH251 CTQYVRHSSD261 PFYTPEPDTC 271 HELLGHVPLL281 AEPSFAQFSQ291 EIGLASLGAS301 EEAVQKLATC311 YFFTVEFGLC 321 KQDGQLRVFG331 AGLLSSISEL341 KHALSGHAKV351 KPFDPKITCK361 QECLITTFQD 371 VYFVSESFED381 AKEKMREFTK391 TIK
|
|||||
|
|
LEU123
4.239
TYR235
3.777
LEU236
3.295
SER237
4.242
PRO238
3.818
PHE241
4.213
LEU242
4.922
ARG257
2.635
PHE263
4.586
TYR264
3.395
THR265
2.859
PRO266
3.126
GLU267
3.532
PRO268
3.615
|
|||||
| Ligand Name: 7,8-dihydrobiopterin | Ligand Info | |||||
| Structure Description | Crystal structure of human tryptophan hydroxylase with bound 7,8-dihydro-L-biopterin cofactor and Fe(III) | PDB:1MLW | ||||
| Method | X-ray diffraction | Resolution | 1.71 Å | Mutation | No | [6] |
| PDB Sequence |
SVPWFPKKIS
113 DLDHCANRVL123 MYGSELDADH133 PGFKDNVYRK143 RRKYFADLAM153 NYKHGDPIPK 163 VEFTEEEIKT173 WGTVFRELNK183 LYPTHACREY193 LKNLPLLSKY203 CGYREDNIPQ 213 LEDVSNFLKE223 RTGFSIRPVA233 GYLSPRDFLS243 GLAFRVFHCT253 QYVRHSSDPF 263 YTPEPDTCHE273 LLGHVPLLAE283 PSFAQFSQEI293 GLASLGASEE303 AVQKLATCYF 313 FTVEFGLCKQ323 DGQLRVFGAG333 LLSSISELKH343 ALSGHAKVKP353 FDPKITCKQE 363 CLITTFQDVY373 FVSESFEDAK383 EKMREFTKTI393
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
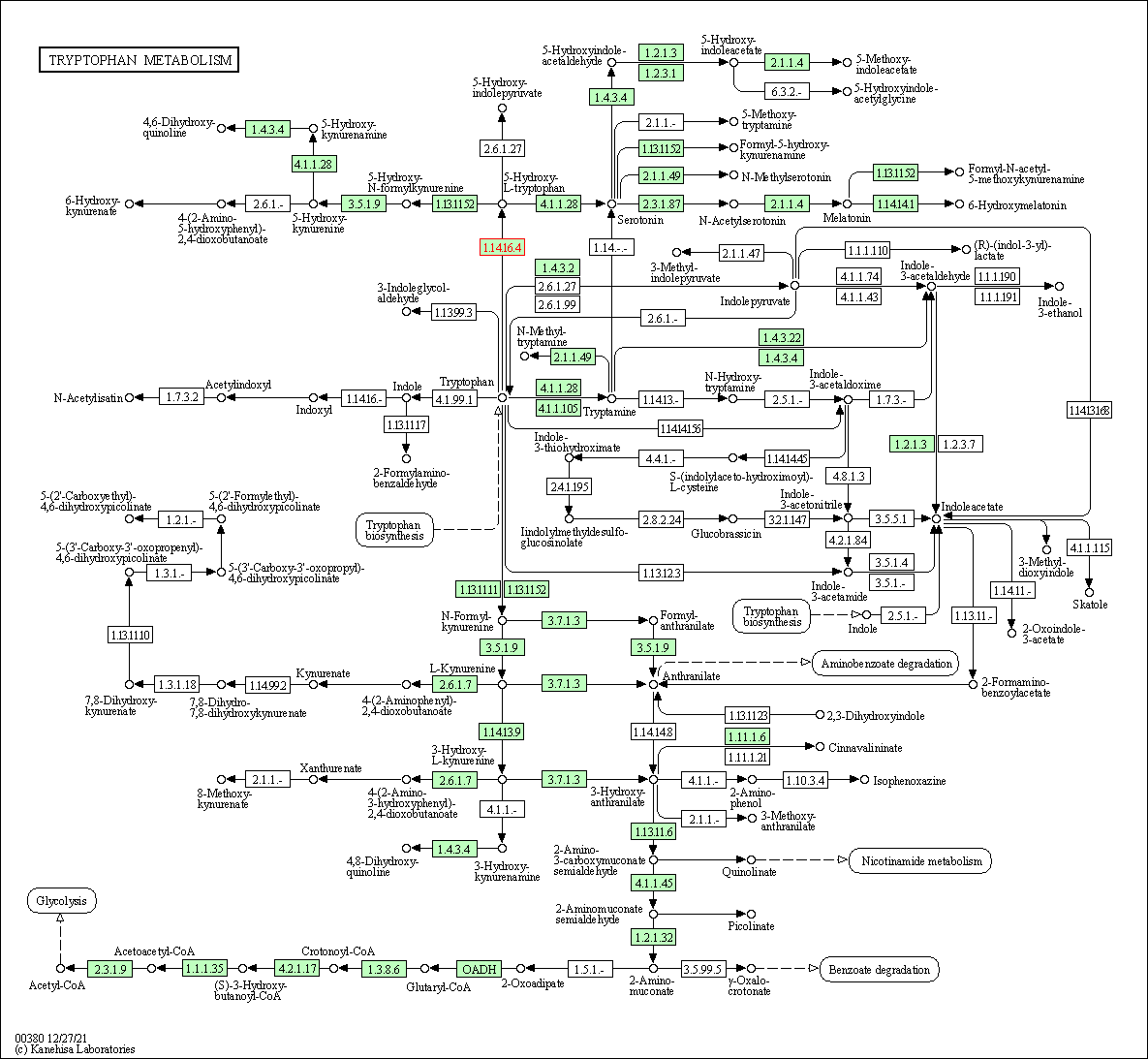
| Tryptophan metabolism | hsa00380 | Affiliated Target |

|
| Class: Metabolism => Amino acid metabolism | Pathway Hierarchy | ||
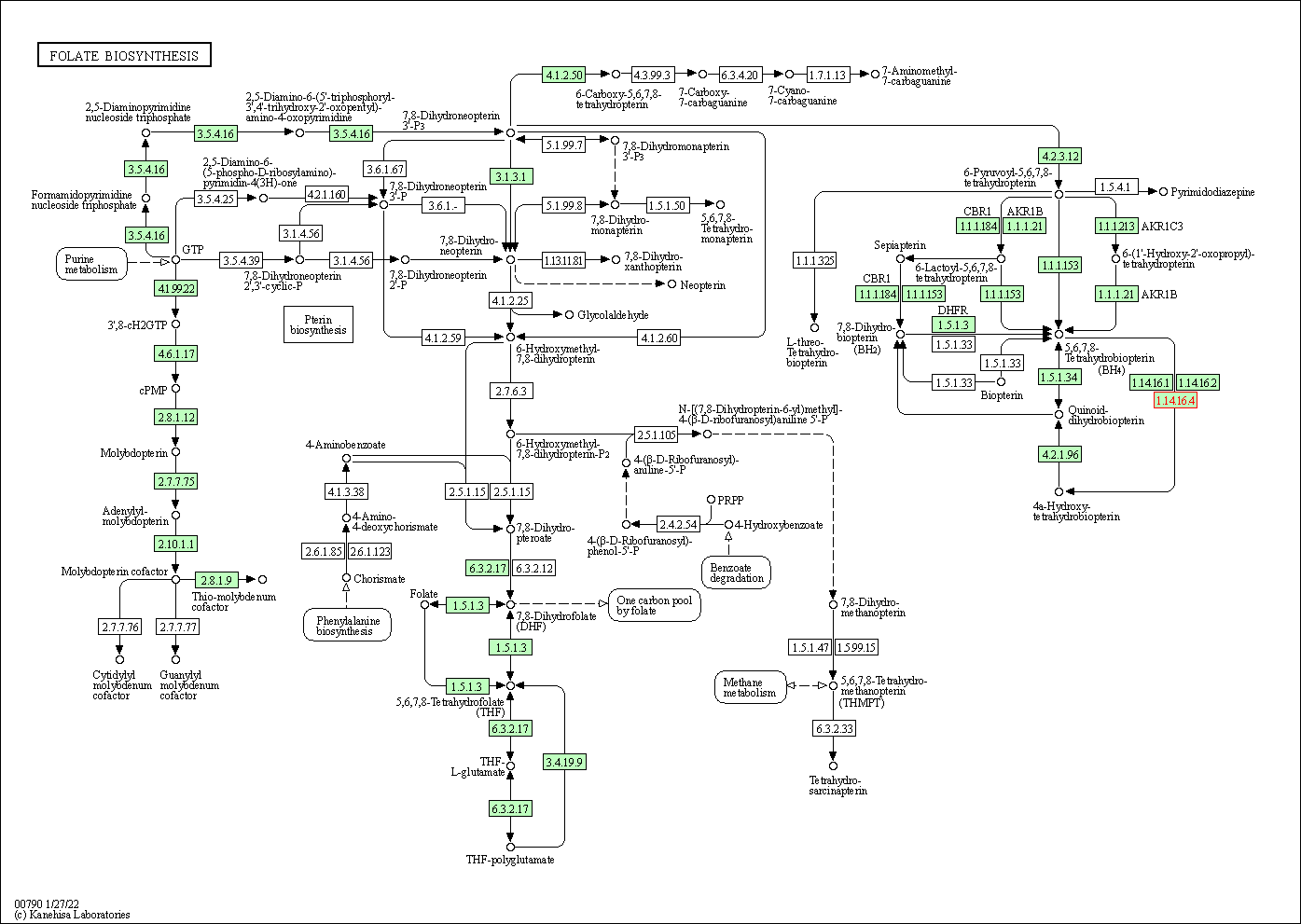
| Folate biosynthesis | hsa00790 | Affiliated Target |

|
| Class: Metabolism => Metabolism of cofactors and vitamins | Pathway Hierarchy | ||
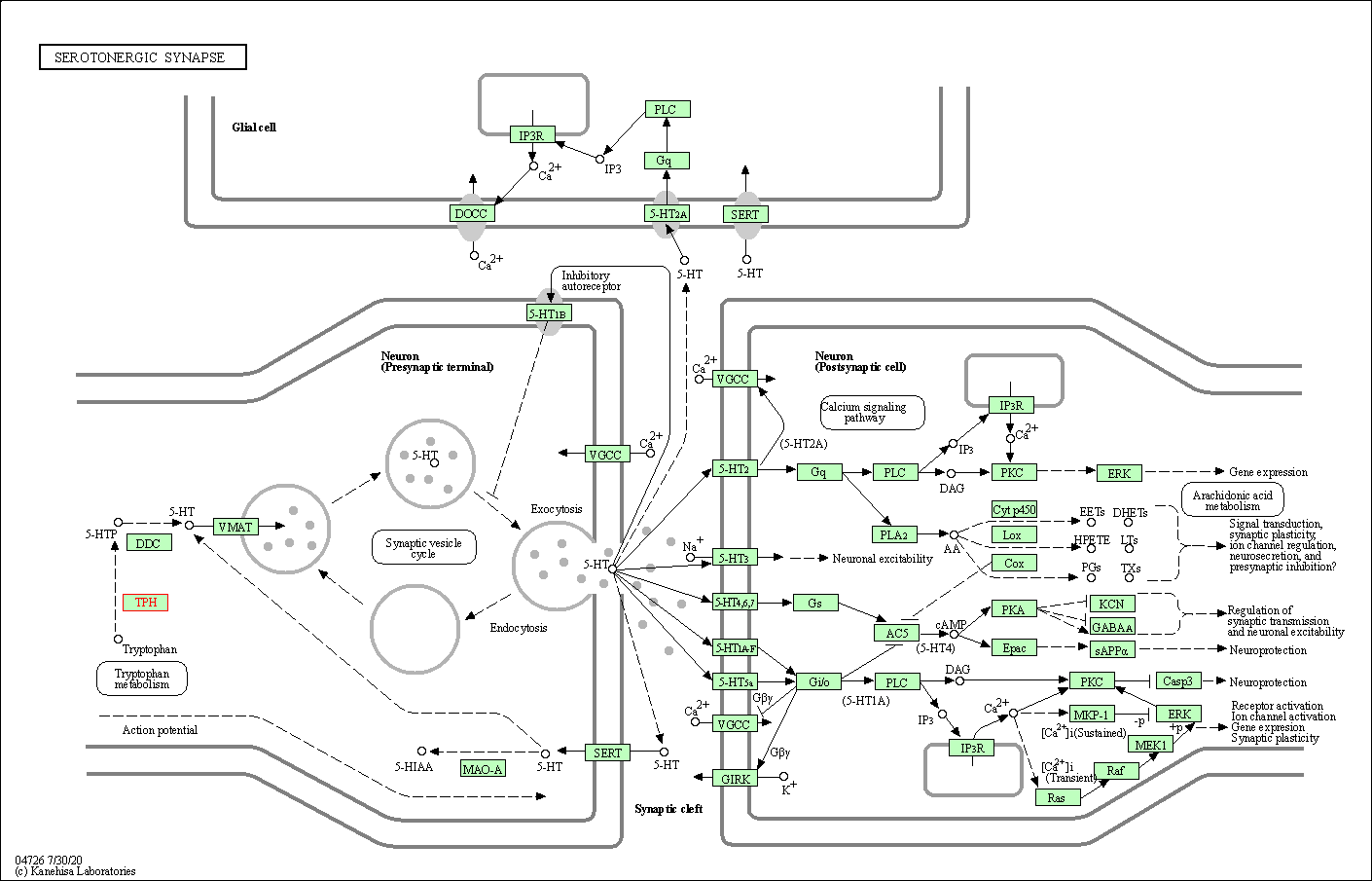
| Serotonergic synapse | hsa04726 | Affiliated Target |

|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Degree | 5 | Degree centrality | 5.37E-04 | Betweenness centrality | 5.14E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 1.65E-01 | Radiality | 1.25E+01 | Clustering coefficient | 2.00E-01 |
| Neighborhood connectivity | 7.40E+00 | Topological coefficient | 2.92E-01 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| BioCyc | [+] 2 BioCyc Pathways | + | ||||
| 1 | Superpathway of tryptophan utilization | |||||
| 2 | Serotonin and melatonin biosynthesis | |||||
| KEGG Pathway | [+] 3 KEGG Pathways | + | ||||
| 1 | Tryptophan metabolism | |||||
| 2 | Metabolic pathways | |||||
| 3 | Serotonergic synapse | |||||
| Panther Pathway | [+] 1 Panther Pathways | + | ||||
| 1 | 5-Hydroxytryptamine biosynthesis | |||||
| Pathwhiz Pathway | [+] 1 Pathwhiz Pathways | + | ||||
| 1 | Tryptophan Metabolism | |||||
| WikiPathways | [+] 4 WikiPathways | + | ||||
| 1 | SIDS Susceptibility Pathways | |||||
| 2 | Biogenic Amine Synthesis | |||||
| 3 | Tryptophan metabolism | |||||
| 4 | Metabolism of amino acids and derivatives | |||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Antidepressant-like action of AGN 2979, a tryptophan hydroxylase activation inhibitor, in a chronic mild stress model of depression in rats. Eur Neuropsychopharmacol. 2001 Oct;11(5):351-7. | |||||
| REF 2 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 3 | (+)-6-fluorotryptophan, an inhibitor of tryptophan hydroxylase: sleep and wakefulness in the rat. Neuropharmacology. 1981 Apr;20(4):335-9. | |||||
| REF 4 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Target id: 1241). | |||||
| REF 5 | Optimization of spirocyclic proline tryptophan hydroxylase-1 inhibitors. Bioorg Med Chem Lett. 2017 Feb 1;27(3):413-419. | |||||
| REF 6 | Three-dimensional structure of human tryptophan hydroxylase and its implications for the biosynthesis of the neurotransmitters serotonin and melatonin. Biochemistry. 2002 Oct 22;41(42):12569-74. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

