Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T77546
(Former ID: TTDR00010)
|
|||||
| Target Name |
Diamine oxidase (AOC1)
|
|||||
| Synonyms |
Kidney amine oxidase; KAO; Histaminase; Amiloride-binding protein; AOC1; ABP
Click to Show/Hide
|
|||||
| Gene Name |
AOC1
|
|||||
| Target Type |
Literature-reported target
|
[1] | ||||
| Function |
Catalyzes the degradation of compounds such as putrescine, histamine, spermine, and spermidine, substances involved in allergic and immune responses, cell proliferation, tissue differentiation, tumor formation, and possibly apoptosis. Placental DAO is thought to play a role in the regulation of the female reproductive function.
Click to Show/Hide
|
|||||
| BioChemical Class |
CH-NH(2) donor oxidoreductase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 1.4.3.22
|
|||||
| Sequence |
MPALGWAVAAILMLQTAMAEPSPGTLPRKAGVFSDLSNQELKAVHSFLWSKKELRLQPSS
TTTMAKNTVFLIEMLLPKKYHVLRFLDKGERHPVREARAVIFFGDQEHPNVTEFAVGPLP GPCYMRALSPRPGYQSSWASRPISTAEYALLYHTLQEATKPLHQFFLNTTGFSFQDCHDR CLAFTDVAPRGVASGQRRSWLIIQRYVEGYFLHPTGLELLVDHGSTDAGHWAVEQVWYNG KFYGSPEELARKYADGEVDVVVLEDPLPGGKGHDSTEEPPLFSSHKPRGDFPSPIHVSGP RLVQPHGPRFRLEGNAVLYGGWSFAFRLRSSSGLQVLNVHFGGERIAYEVSVQEAVALYG GHTPAGMQTKYLDVGWGLGSVTHELAPGIDCPETATFLDTFHYYDADDPVHYPRALCLFE MPTGVPLRRHFNSNFKGGFNFYAGLKGQVLVLRTTSTVYNYDYIWDFIFYPNGVMEAKMH ATGYVHATFYTPEGLRHGTRLHTHLIGNIHTHLVHYRVDLDVAGTKNSFQTLQMKLENIT NPWSPRHRVVQPTLEQTQYSWERQAAFRFKRKLPKYLLFTSPQENPWGHKRTYRLQIHSM ADQVLPPGWQEEQAITWARYPLAVTKYRESELCSSSIYHQNDPWHPPVVFEQFLHNNENI ENEDLVAWVTVGFLHIPHSEDIPNTATPGNSVGFLLRPFNFFPEDPSLASRDTVIVWPRD NGPNYVQRWIPEDRDCSMPPPFSYNGTYRPV Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Pentamidine | Ligand Info | |||||
| Structure Description | Crystal structure of human diamine oxidase in complex with the inhibitor pentamidine | PDB:3HII | ||||
| Method | X-ray diffraction | Resolution | 2.15 Å | Mutation | Yes | [2] |
| PDB Sequence |
PRKAGVFSDL
36 SNQELKAVHS46 FLWSKKELRL56 QPSSTTTMAK66 NTVFLIEMLL76 PKKYHVLRFL 86 DKGERHPVRE96 ARAVIFFGDQ106 EHPNVTEFAV116 GPLPGPCYMR126 ALSPRPGYQS 136 SWASRPISTA146 EYALLYHTLQ156 EATKPLHQFF166 LNTTGFSFQD176 CHDRCLAFTD 186 VAPRGVASGQ196 RRSWLIIQRY206 VEGYFLHPTG216 LELLVDHGST226 DAGHWAVEQV 236 WYNGKFYGSP246 EELARKYADG256 EVDVVVLEDP266 LEPPLFSSHK286 PRGDFPSPIH 296 VSGPRLVQPH306 GPRFRLEGNA316 VLYGGWSFAF326 RLRSSSGLQV336 LNVHFGGERI 346 AYEVSVQEAV356 ALYGGHTPAG366 MQTKYLDVGW376 GLGSVTHELA386 PGIDCPETAT 396 FLDTFHYYDA406 DDPVHYPRAL416 CLFEMPTGVP426 LRRHFNSNFK436 GGFNFYAGLK 446 GQVLVLRTTS456 TVYNDYIWDF467 IFYPNGVMEA477 KMHATGYVHA487 TFYTPEGLRH 497 GTRLHTHLIG507 NIHTHLVHYR517 VDLDVAGTKN527 SFQTLQMKLE537 NITNPWSPRH 547 RVVQPTLEQT557 QYSWERQAAF567 RFKRKLPKYL577 LFTSPQENPW587 GHKRSYRLQI 597 HSMADQVLPP607 GWQEEQAITW617 ARYPLAVTKY627 RESELCSSSI637 YHQNDPWDPP 647 VVFEQFLHNN657 ENIENEDLVA667 WVTVGFLHIP677 HSEDIPNTAT687 PGNSVGFLLR 697 PFNFFPEDPS707 LASRDTVIVW717 PRDNGPNYVQ727 RWIPEDRDCS737 MPPPFSYNGT 747 YRPV
|
|||||
|
|
||||||
| Ligand Name: 6-hydroxydopa quinone | Ligand Info | |||||
| Structure Description | Crystal structure of human diamine oxidase in complex with the inhibitor berenil | PDB:3HIG | ||||
| Method | X-ray diffraction | Resolution | 2.09 Å | Mutation | Yes | [2] |
| PDB Sequence |
PRKAGVFSDL
36 SNQELKAVHS46 FLWSKKELRL56 QPSSTTTMAK66 NTVFLIEMLL76 PKKYHVLRFL 86 DKGERHPVRE96 ARAVIFFGDQ106 EHPNVTEFAV116 GPLPGPCYMR126 ALSPRPGYQS 136 SWASRPISTA146 EYALLYHTLQ156 EATKPLHQFF166 LNTTGFSFQD176 CHDRCLAFTD 186 VAPRGVASGQ196 RRSWLIIQRY206 VEGYFLHPTG216 LELLVDHGST226 DAGHWAVEQV 236 WYNGKFYGSP246 EELARKYADG256 EVDVVVLEDP266 LEPPLFSSHK286 PRGDFPSPIH 296 VSGPRLVQPH306 GPRFRLEGNA316 VLYGGWSFAF326 RLRSSSGLQV336 LNVHFGGERI 346 AYEVSVQEAV356 ALYGGHTPAG366 MQTKYLDVGW376 GLGSVTHELA386 PGIDCPETAT 396 FLDTFHYYDA406 DDPVHYPRAL416 CLFEMPTGVP426 LRRHFNSNFK436 GGFNFYAGLK 446 GQVLVLRTTS456 TVYNDYIWDF467 IFYPNGVMEA477 KMHATGYVHA487 TFYTPEGLRH 497 GTRLHTHLIG507 NIHTHLVHYR517 VDLDVAGTKN527 SFQTLQMKLE537 NITNPWSPRH 547 RVVQPTLEQT557 QYSWERQAAF567 RFKRKLPKYL577 LFTSPQENPW587 GHKRSYRLQI 597 HSMADQVLPP607 GWQEEQAITW617 ARYPLAVTKY627 RESELCSSSI637 YHQNDPWDPP 647 VVFEQFLHNN657 ENIENEDLVA667 WVTVGFLHIP677 HSEDIPNTAT687 PGNSVGFLLR 697 PFNFFPEDPS707 LASRDTVIVW717 PRDNGPNYVQ727 RWIPEDRDCS737 MPPPFSYNGT 747 YRPV
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
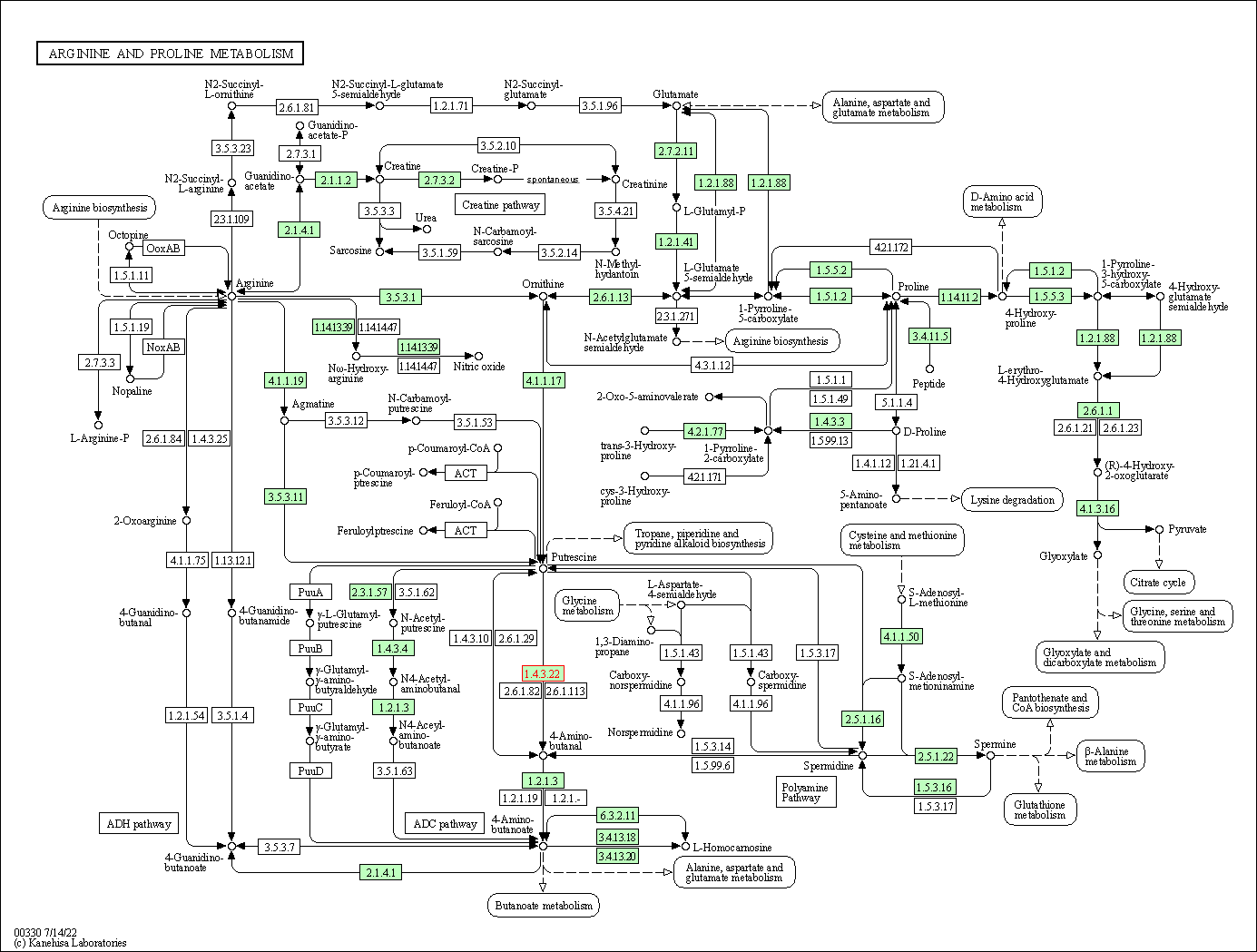
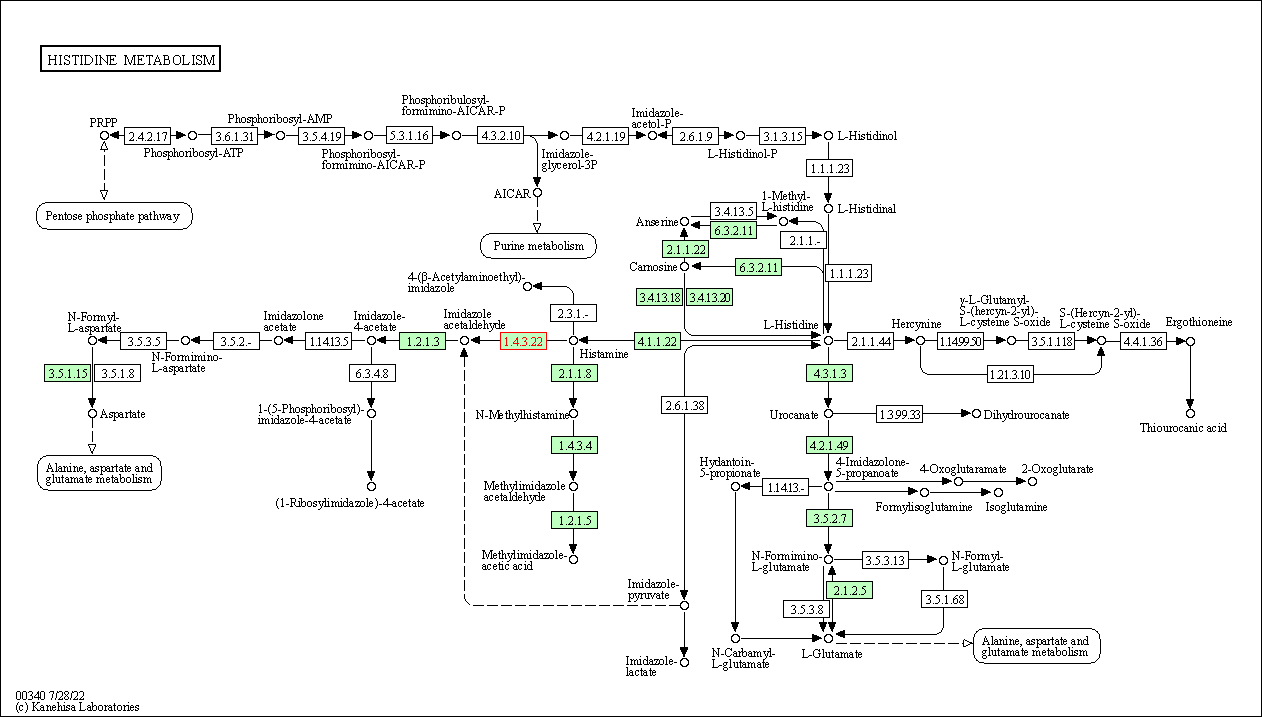
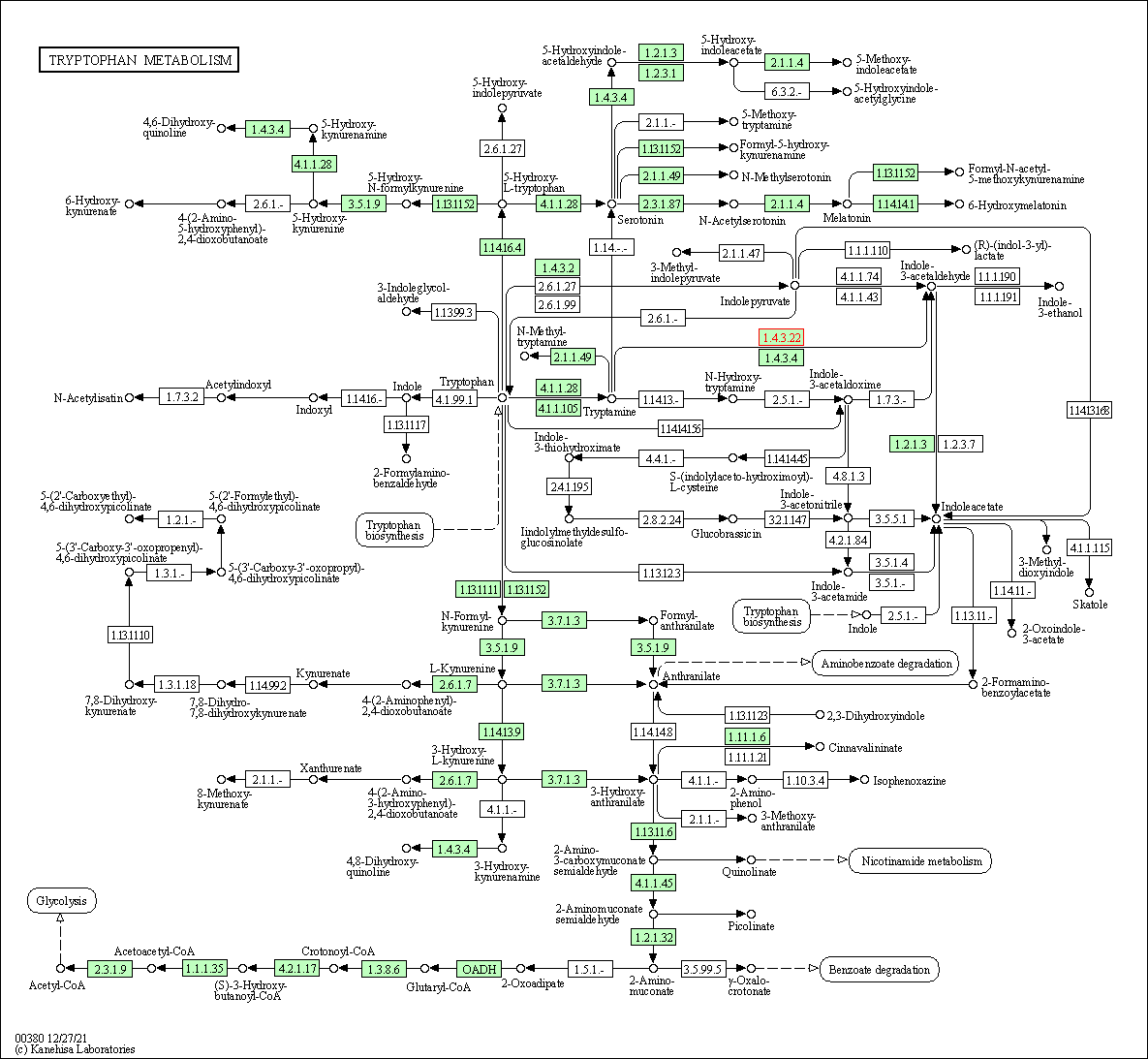
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Arginine and proline metabolism | hsa00330 | Affiliated Target |

|
| Class: Metabolism => Amino acid metabolism | Pathway Hierarchy | ||
| Histidine metabolism | hsa00340 | Affiliated Target |

|
| Class: Metabolism => Amino acid metabolism | Pathway Hierarchy | ||
| Tryptophan metabolism | hsa00380 | Affiliated Target |

|
| Class: Metabolism => Amino acid metabolism | Pathway Hierarchy | ||
| Degree | 5 | Degree centrality | 5.37E-04 | Betweenness centrality | 1.54E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 1.67E-01 | Radiality | 1.26E+01 | Clustering coefficient | 2.00E-01 |
| Neighborhood connectivity | 6.60E+00 | Topological coefficient | 2.56E-01 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| BioCyc | [+] 1 BioCyc Pathways | + | ||||
| 1 | Histamine degradation | |||||
| KEGG Pathway | [+] 3 KEGG Pathways | + | ||||
| 1 | Arginine and proline metabolism | |||||
| 2 | Histidine metabolism | |||||
| 3 | Tryptophan metabolism | |||||
| Panther Pathway | [+] 1 Panther Pathways | + | ||||
| 1 | Phenylethylamine degradation | |||||
| Pathwhiz Pathway | [+] 2 Pathwhiz Pathways | + | ||||
| 1 | Histidine Metabolism | |||||
| 2 | Tyrosine Metabolism | |||||
| WikiPathways | [+] 1 WikiPathways | + | ||||
| 1 | Tryptophan metabolism | |||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | How many drug targets are there Nat Rev Drug Discov. 2006 Dec;5(12):993-6. | |||||
| REF 2 | Structure and inhibition of human diamine oxidase. Biochemistry. 2009 Oct 20;48(41):9810-22. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

