Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T67272
(Former ID: TTDR00784)
|
|||||
| Target Name |
Aminopeptidase N (ANPEP)
|
|||||
| Synonyms |
Myeloid plasma membrane glycoprotein CD13; Microsomal aminopeptidase; HAPN; Gp150; Aminopeptidase M; Alanyl aminopeptidase; ANPEP
Click to Show/Hide
|
|||||
| Gene Name |
ANPEP
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Psoriasis [ICD-11: EA90] | |||||
| Function |
Broad specificity aminopeptidase. Plays a role in the final digestion of peptides generated from hydrolysis of proteins by gastric and pancreatic proteases. May play a critical role in the pathogenesis of cholesterol gallstone disease. May be involved in the metabolism of regulatory peptides of diverse cell types, responsible for the processing of peptide hormones, such as angiotensin III and IV, neuropeptides, and chemokines. Found to cleave antigen peptides bound to major histocompatibility complex class II molecules of presenting cells and to degrade neurotransmitters at synaptic junctions. Is also implicated as a regulator of IL-8 bioavailability in the endometrium, and therefore may contribute to the regulation of angiogenesis. Is used as a marker for acute myeloid leukemia and plays a role in tumor invasion. In case of human coronavirus 229E (HCoV-229E) infection, serves as receptor for HCoV-229E spike glycoprotein. Mediates as well human cytomegalovirus (HCMV) infection.
Click to Show/Hide
|
|||||
| BioChemical Class |
Peptidase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 3.4.11.2
|
|||||
| Sequence |
MAKGFYISKSLGILGILLGVAAVCTIIALSVVYSQEKNKNANSSPVASTTPSASATTNPA
SATTLDQSKAWNRYRLPNTLKPDSYRVTLRPYLTPNDRGLYVFKGSSTVRFTCKEATDVI IIHSKKLNYTLSQGHRVVLRGVGGSQPPDIDKTELVEPTEYLVVHLKGSLVKDSQYEMDS EFEGELADDLAGFYRSEYMEGNVRKVVATTQMQAADARKSFPCFDEPAMKAEFNITLIHP KDLTALSNMLPKGPSTPLPEDPNWNVTEFHTTPKMSTYLLAFIVSEFDYVEKQASNGVLI RIWARPSAIAAGHGDYALNVTGPILNFFAGHYDTPYPLPKSDQIGLPDFNAGAMENWGLV TYRENSLLFDPLSSSSSNKERVVTVIAHELAHQWFGNLVTIEWWNDLWLNEGFASYVEYL GADYAEPTWNLKDLMVLNDVYRVMAVDALASSHPLSTPASEINTPAQISELFDAISYSKG ASVLRMLSSFLSEDVFKQGLASYLHTFAYQNTIYLNLWDHLQEAVNNRSIQLPTTVRDIM NRWTLQMGFPVITVDTSTGTLSQEHFLLDPDSNVTRPSEFNYVWIVPITSIRDGRQQQDY WLIDVRAQNDLFSTSGNEWVLLNLNVTGYYRVNYDEENWRKIQTQLQRDHSAIPVINRAQ IINDAFNLASAHKVPVTLALNNTLFLIEERQYMPWEAALSSLSYFKLMFDRSEVYGPMKN YLKKQVTPLFIHFRNNTNNWREIPENLMDQYSEVNAISTACSNGVPECEEMVSGLFKQWM ENPNNNPIHPNLRSTVYCNAIAQGGEEEWDFAWEQFRNATLVNEADKLRAALACSKELWI LNRYLSYTLNPDLIRKQDATSTIISITNNVIGQGLVWDFVQSNWKKLFNDYGGGSFSFSN LIQAVTRRFSTEYELQQLEQFKKDNEETGFGSGTRALEQALEKTKANIKWVKENKEVVLQ WFTENSK Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| HIT2.0 ID | T64E3A | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 1 Clinical Trial Drugs | + | ||||
| 1 | IP10 C8 | Drug Info | Phase 2 | Psoriasis vulgaris | [1] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Modulator | [+] 1 Modulator drugs | + | ||||
| 1 | IP10 C8 | Drug Info | [1] | |||
| Inhibitor | [+] 39 Inhibitor drugs | + | ||||
| 1 | (1-Amino-2-phenyl-ethyl)-phosphinic acid | Drug Info | [2] | |||
| 2 | (1-Amino-3-methyl-butyl)-phosphinic acid | Drug Info | [2] | |||
| 3 | (1-Amino-3-methylsulfanyl-propyl)-phosphinic acid | Drug Info | [2] | |||
| 4 | (1-Amino-3-phenyl-propyl)-phosphinic acid | Drug Info | [2] | |||
| 5 | (1-Amino-ethyl)-phosphinic acid | Drug Info | [2] | |||
| 6 | (Amino-phenyl-methyl)-phosphinic acid | Drug Info | [2] | |||
| 7 | (S)-2-Amino-2-phenyl-ethanethiol | Drug Info | [2] | |||
| 8 | (S)-2-Amino-3-phenyl-propane-1-thiol | Drug Info | [2] | |||
| 9 | (S)-2-Amino-4-methyl-pentane-1-thiol | Drug Info | [2] | |||
| 10 | (S)-2-Amino-4-methylsulfanyl-butane-1-thiol | Drug Info | [2] | |||
| 11 | (S)-2-Amino-4-phenyl-butane-1-thiol | Drug Info | [2] | |||
| 12 | (S)-2-Amino-propane-1-thiol | Drug Info | [2] | |||
| 13 | 1-aminobutylphosphonic acid | Drug Info | [3] | |||
| 14 | 1-aminohexylphosphonic acid | Drug Info | [3] | |||
| 15 | 1-aminohexylphosphonic acid monophenyl ester | Drug Info | [3] | |||
| 16 | 1-aminopentylphosphonic acid monophenyl ester | Drug Info | [3] | |||
| 17 | 2-amino-N1-benzyl-N3-hydroxymalonamide | Drug Info | [4] | |||
| 18 | 2-benzyl-N1-hydroxy-N3-(3-phenylpropyl)malonamide | Drug Info | [4] | |||
| 19 | 2-benzyl-N1-hydroxy-N3-(4-phenylbutyl)malonamide | Drug Info | [4] | |||
| 20 | 2-benzyl-N1-hydroxy-N3-phenethylmalonamide | Drug Info | [4] | |||
| 21 | 2-Cinnamamido-N1-hydroxy-N4-octylsuccinamide | Drug Info | [5] | |||
| 22 | 2-Cinnamamido-N1-hydroxy-N4-pentylsuccinamide | Drug Info | [5] | |||
| 23 | 2-Cinnamamido-N4-hexyl-N1-hydroxysuccinamide | Drug Info | [5] | |||
| 24 | Angiotensin IV | Drug Info | [6] | |||
| 25 | KELATORPHAN | Drug Info | [7] | |||
| 26 | N-biphenyl-3-ylmethyl-N'-hydroxy-malonamide | Drug Info | [4] | |||
| 27 | N-Hydroxy-2-(naphthalen-2-ylsulfanyl)-acetamide | Drug Info | [8] | |||
| 28 | N1,2-dibenzyl-N3-hydroxy-N1-phenethylmalonamide | Drug Info | [4] | |||
| 29 | N1,2-dibenzyl-N3-hydroxymalonamide | Drug Info | [4] | |||
| 30 | N1-(3,3-diphenylpropyl)-N3-hydroxymalonamide | Drug Info | [4] | |||
| 31 | N1-(3-phenoxybenzyl)-N3-hydroxymalonamide | Drug Info | [4] | |||
| 32 | N1-(4-chlorobenzyl)-2-amino-N3-hydroxymalonamide | Drug Info | [4] | |||
| 33 | N1-(4-chlorobenzyl)-2-benzyl-N3-hydroxymalonamide | Drug Info | [4] | |||
| 34 | N1-(4-fluorobenzyl)-2-benzyl-N3-hydroxymalonamide | Drug Info | [4] | |||
| 35 | N1-benzyl-N3-hydroxy-2-isobutylmalonamide | Drug Info | [9] | |||
| 36 | N1-hydroxy-2-isobutyl-N3-phenethylmalonamide | Drug Info | [9] | |||
| 37 | N4-Butyl-2-cinnamamido-N1-hydroxysuccinamide | Drug Info | [5] | |||
| 38 | PMID23428964CI3 | Drug Info | [10] | |||
| 39 | [(125)I] RB129 | Drug Info | [11] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Bestatin | Ligand Info | |||||
| Structure Description | Human aminopeptidase N (CD13) in complex with bestatin | PDB:4FYR | ||||
| Method | X-ray diffraction | Resolution | 1.91 Å | Mutation | No | [12] |
| PDB Sequence |
PDQSKAWNRY
74 RLPNTLKPDS84 YRVTLRPYLT94 PNDRGLYVFK104 GSSTVRFTCK114 EATDVIIIHS 124 KKLNYTLSQG134 HRVVLRGVGG144 SQPPDIDKTE154 LVEPTEYLVV164 HLKGSLVKDS 174 QYEMDSEFEG184 ELADDLAGFY194 RSEYMEGNVR204 KVVATTQMQA214 ADARKSFPCF 224 DEPAMKAEFN234 ITLIHPKDLT244 ALSNMLPKGP254 STPLPEDPNW264 NVTEFHTTPK 274 MSTYLLAFIV284 SEFDYVEKQA294 SNGVLIRIWA304 RPSAIAAGHG314 DYALNVTGPI 324 LNFFAGHYDT334 PYPLPKSDQI344 GLPDFNAGAM354 ENWGLVTYRE364 NSLLFDPLSS 374 SSSNKERVVT384 VIAHELAHQW394 FGNLVTIEWW404 NDLWLNEGFA414 SYVEYLGADY 424 AEPTWNLKDL434 MVLNDVYRVM444 AVDALASSHP454 LSTPASEINT464 PAQISELFDA 474 ISYSKGASVL484 RMLSSFLSED494 VFKQGLASYL504 HTFAYQNTIY514 LNLWDHLQEA 524 VNNRSIQLPT534 TVRDIMNRWT544 LQMGFPVITV554 DTSTGTLSQE564 HFLLDPDSNV 574 TRPSEFNYVW584 IVPITSIRDG594 RQQQDYWLID604 VRAQNDLFST614 SGNEWVLLNL 624 NVTGYYRVNY634 DEENWRKIQT644 QLQRDHSAIP654 VINRAQIIND664 AFNLASAHKV 674 PVTLALNNTL684 FLIEERQYMP694 WEAALSSLSY704 FKLMFDRSEV714 YGPMKNYLKK 724 QVTPLFIHFR734 NNTNNWREIP744 ENLMDQYSEV754 NAISTACSNG764 VPECEEMVSG 774 LFKQWMENPN784 NNPIHPNLRS794 TVYCNAIAQG804 GEEEWDFAWE814 QFRNATLVNE 824 ADKLRAALAC834 SKELWILNRY844 LSYTLNPDLI854 RKQDATSTII864 SITNNVIGQG 874 LVWDFVQSNW884 KKLFNDYGGG894 SFSFSNLIQA904 VTRRFSTEYE914 LQQLEQFKKD 924 NEETGFGSGT934 RALEQALEKT944 KANIKWVKEN954 KEVVLQWFTE964 NSK |
|||||
|
|
GLN211
4.014
GLN213
3.108
ALA214
3.681
ALA351
3.608
GLY352
4.356
ALA353
3.159
MET354
4.302
GLU355
3.328
ARG381
4.449
VAL385
3.981
|
|||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Glutathione metabolism | hsa00480 | Affiliated Target |
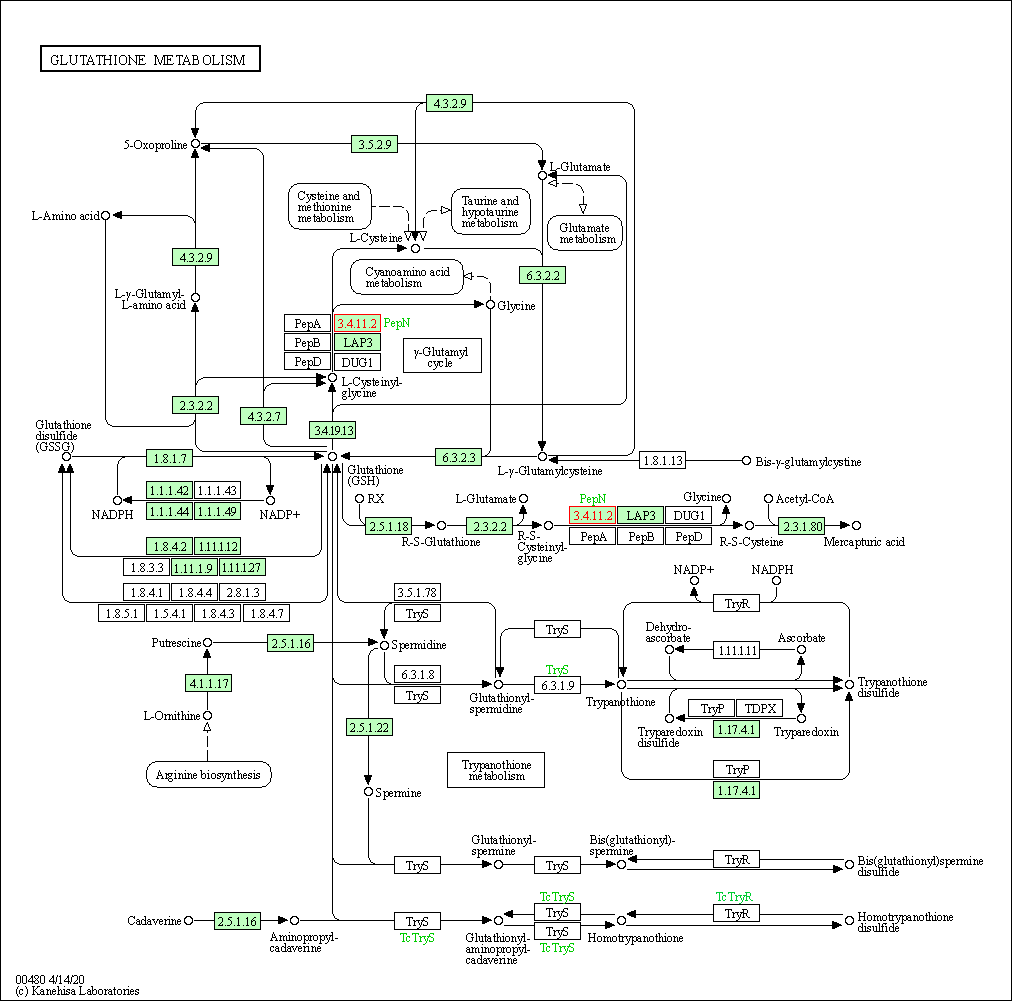
|
| Class: Metabolism => Metabolism of other amino acids | Pathway Hierarchy | ||
| Renin-angiotensin system | hsa04614 | Affiliated Target |
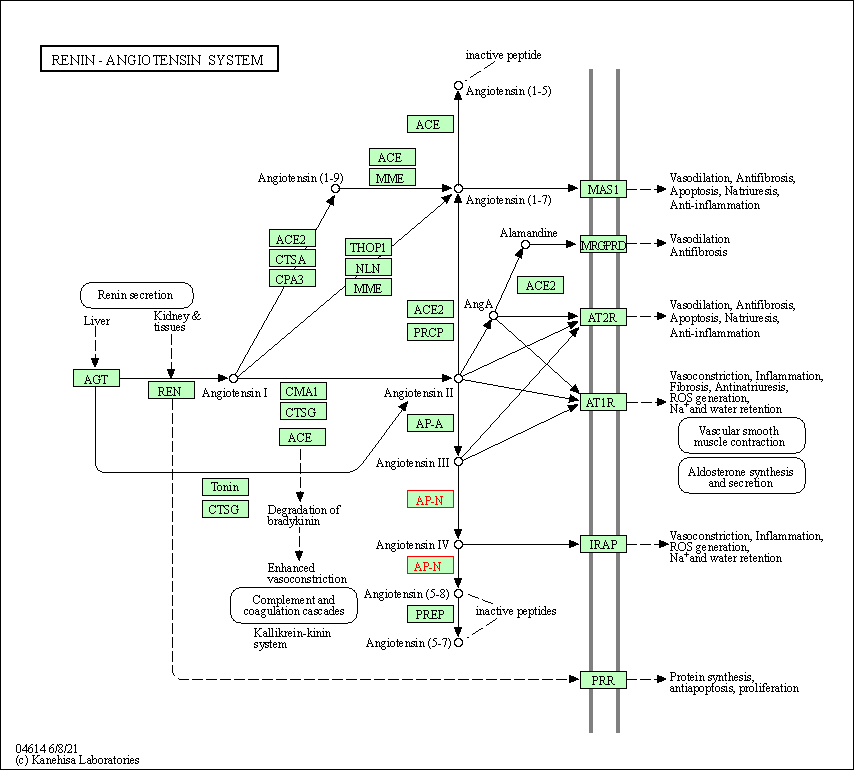
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Hematopoietic cell lineage | hsa04640 | Affiliated Target |
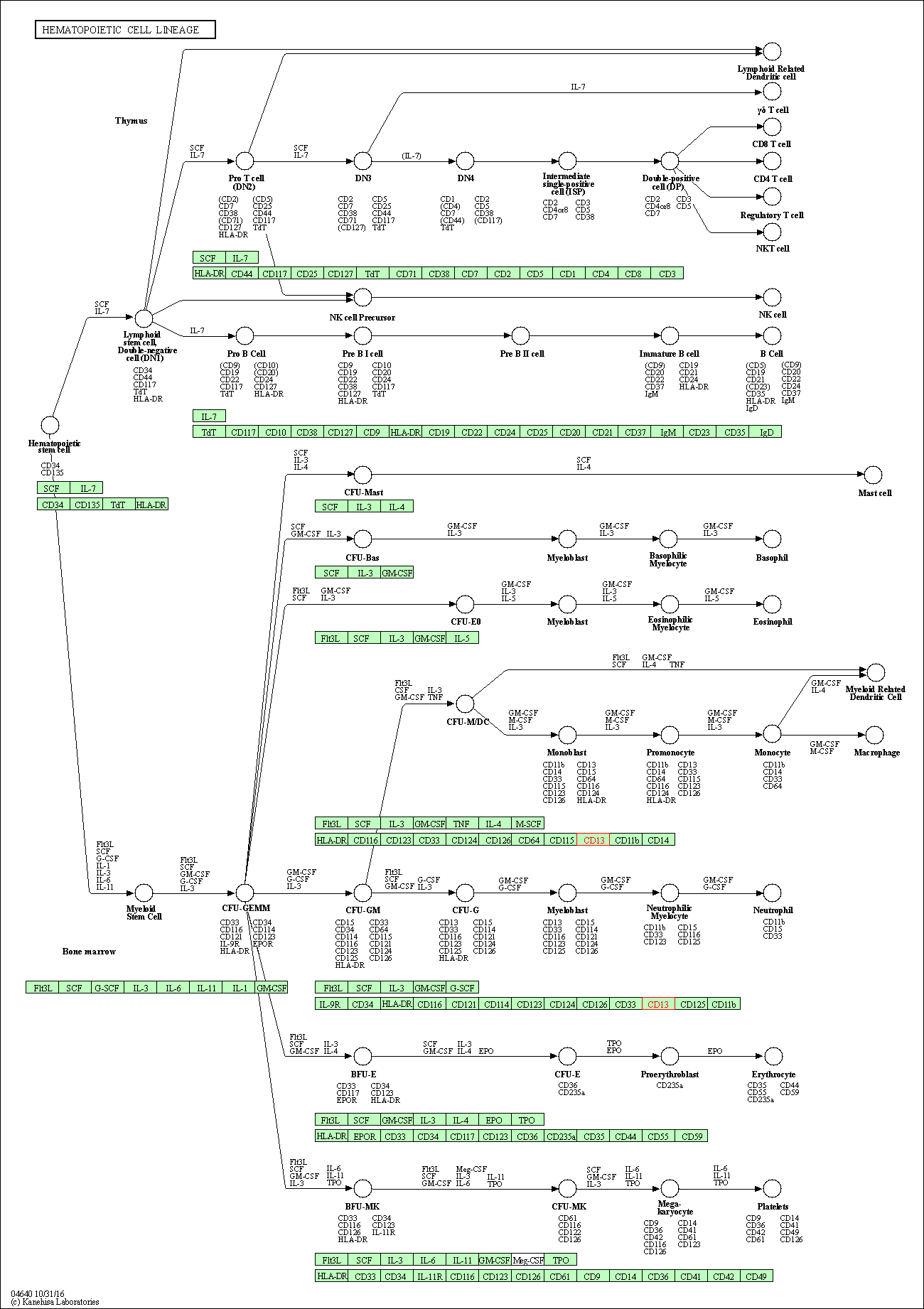
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Degree | 4 | Degree centrality | 4.30E-04 | Betweenness centrality | 2.09E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 2.10E-01 | Radiality | 1.37E+01 | Clustering coefficient | 0.00E+00 |
| Neighborhood connectivity | 1.35E+01 | Topological coefficient | 2.55E-01 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| BioCyc | [+] 1 BioCyc Pathways | + | ||||
| 1 | Glutathione-mediated detoxification | |||||
| KEGG Pathway | [+] 4 KEGG Pathways | + | ||||
| 1 | Glutathione metabolism | |||||
| 2 | Metabolic pathways | |||||
| 3 | Renin-angiotensin system | |||||
| 4 | Hematopoietic cell lineage | |||||
| Pathwhiz Pathway | [+] 1 Pathwhiz Pathways | + | ||||
| 1 | Glutathione Metabolism | |||||
| PID Pathway | [+] 1 PID Pathways | + | ||||
| 1 | C-MYB transcription factor network | |||||
| Reactome | [+] 1 Reactome Pathways | + | ||||
| 1 | Metabolism of Angiotensinogen to Angiotensins | |||||
| WikiPathways | [+] 7 WikiPathways | + | ||||
| 1 | Metabolism of Angiotensinogen to Angiotensins | |||||
| 2 | Cardiac Progenitor Differentiation | |||||
| 3 | miR-targeted genes in squamous cell - TarBase | |||||
| 4 | miR-targeted genes in muscle cell - TarBase | |||||
| 5 | miR-targeted genes in lymphocytes - TarBase | |||||
| 6 | miR-targeted genes in leukocytes - TarBase | |||||
| 7 | Glutathione metabolism | |||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Recent insights into the role of dipeptidyl aminopeptidase IV (DPIV) and aminopeptidase N (APN) families in immune functions. Clin Chem Lab Med. 2009;47(3):253-61. | |||||
| REF 2 | Design of the first highly potent and selective aminopeptidase N (EC 3.4.11.2) inhibitor. Bioorg Med Chem Lett. 1999 Jun 7;9(11):1511-6. | |||||
| REF 3 | New aromatic monoesters of alpha-aminoaralkylphosphonic acids as inhibitors of aminopeptidase N/CD13. Bioorg Med Chem. 2010 Apr 15;18(8):2930-6. | |||||
| REF 4 | Novel selective inhibitors of the zinc plasmodial aminopeptidase PfA-M1 as potential antimalarial agents. J Med Chem. 2007 Mar 22;50(6):1322-34. | |||||
| REF 5 | Design, synthesis, and preliminary studies of the activity of novel derivatives of N-cinnamoyl-L-aspartic acid as inhibitors of aminopeptidase N/CD13. Bioorg Med Chem. 2009 Oct 15;17(20):7398-404. | |||||
| REF 6 | Ligands to the (IRAP)/AT4 receptor encompassing a 4-hydroxydiphenylmethane scaffold replacing Tyr2. Bioorg Med Chem. 2008 Jul 15;16(14):6924-35. | |||||
| REF 7 | Retro-inverso concept applied to the complete inhibitors of enkephalin-degrading enzymes. J Med Chem. 1988 Sep;31(9):1825-31. | |||||
| REF 8 | N-hydroxy-2-(naphthalene-2-ylsulfanyl)-acetamide, a novel hydroxamic acid-based inhibitor of aminopeptidase N and its anti-angiogenic activity. Bioorg Med Chem Lett. 2005 Jan 3;15(1):181-3. | |||||
| REF 9 | A library of novel hydroxamic acids targeting the metallo-protease family: design, parallel synthesis and screening. Bioorg Med Chem. 2007 Jan 1;15(1):63-76. | |||||
| REF 10 | Selective aminopeptidase-N (CD13) inhibitors with relevance to cancer chemotherapy. Bioorg Med Chem. 2013 Apr 1;21(7):2135-44. | |||||
| REF 11 | Ontogenic and adult whole body distribution of aminopeptidase N in rat investigated by in vitro autoradiography. Biochimie. 2004 Feb;86(2):105-13. | |||||
| REF 12 | The X-ray crystal structure of human aminopeptidase N reveals a novel dimer and the basis for peptide processing. J Biol Chem. 2012 Oct 26;287(44):36804-13. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

