Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T65864
(Former ID: TTDC00201)
|
|||||
| Target Name |
Stress-activated protein kinase 2a (p38 alpha)
|
|||||
| Synonyms |
SAPK2A; P38 mitogen activatedprotein kinase; P38 Mitogen-activatedprotein kinase alpha; Mitogen-activated protein kinase p38 alpha; Mitogen-activated protein kinase 14; MXI2; MAX-interacting protein 2; MAPK 14; MAP kinase p38alpha; MAP kinase p38 alpha; MAP kinase MXI2; MAP kinase 14; Cytokine suppressive anti-inflammatory drug-binding protein; Cytokine suppressive anti-inflammatory drug binding protein; CSPB1; CSBP2; CSBP1; CSBP; CSAID-binding protein; CSAID binding protein; CRK1
Click to Show/Hide
|
|||||
| Gene Name |
MAPK14
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 4 Target-related Diseases | + | ||||
| 1 | Vitamin deficiency [ICD-11: 5B55-5B5F] | |||||
| 2 | Myocardial infarction [ICD-11: BA41-BA43] | |||||
| 3 | Coronary atherosclerosis [ICD-11: BA80] | |||||
| 4 | Rheumatoid arthritis [ICD-11: FA20] | |||||
| Function |
MAPK14 is one of the four p38 MAPKs which play an important role in the cascades of cellular responses evoked by extracellular stimuli such as proinflammatory cytokines or physical stress leading to direct activation of transcription factors. Accordingly, p38 MAPKs phosphorylate a broad range of proteins and it has been estimated that they may have approximately 200 to 300 substrates each. Some of the targets are downstream kinases which are activated through phosphorylation and further phosphorylate additional targets. RPS6KA5/MSK1 and RPS6KA4/MSK2 can directly phosphorylate and activate transcription factors such as CREB1, ATF1, the NF-kappa-B isoform RELA/NFKB3, STAT1 and STAT3, but can also phosphorylate histone H3 and the nucleosomal protein HMGN1. RPS6KA5/MSK1 and RPS6KA4/MSK2 play important roles in the rapid induction of immediate-early genes in response to stress or mitogenic stimuli, either by inducing chromatin remodeling or by recruiting the transcription machinery. On the other hand, two other kinase targets, MAPKAPK2/MK2 and MAPKAPK3/MK3, participate in the control of gene expression mostly at the post-transcriptional level, by phosphorylating ZFP36 (tristetraprolin) and ELAVL1, and by regulating EEF2K, which is important for the elongation of mRNA during translation. MKNK1/MNK1 and MKNK2/MNK2, two other kinases activated by p38 MAPKs, regulate protein synthesis by phosphorylating the initiation factor EIF4E2. MAPK14 interacts also with casein kinase II, leading to its activation through autophosphorylation and further phosphorylation of TP53/p53. In the cytoplasm, the p38 MAPK pathway is an important regulator of protein turnover. For example, CFLAR is an inhibitor of TNF-induced apoptosis whose proteasome-mediated degradation is regulated by p38 MAPK phosphorylation. In a similar way, MAPK14 phosphorylates the ubiquitin ligase SIAH2, regulating its activity towards EGLN3. MAPK14 may also inhibit the lysosomal degradation pathway of autophagy by interfering with the intracellular trafficking of the transmembrane protein ATG9. Another function of MAPK14 is to regulate the endocytosis of membrane receptors by different mechanisms that impinge on the small GTPase RAB5A. In addition, clathrin-mediated EGFR internalization induced by inflammatory cytokines and UV irradiation depends on MAPK14-mediated phosphorylation of EGFR itself as well as of RAB5A effectors. Ectodomain shedding of transmembrane proteins is regulated by p38 MAPKs as well. In response to inflammatory stimuli, p38 MAPKs phosphorylate the membrane-associated metalloprotease ADAM17. Such phosphorylation is required for ADAM17-mediated ectodomain shedding of TGF-alpha family ligands, which results in the activation of EGFR signaling and cell proliferation. Another p38 MAPK substrate is FGFR1. FGFR1 can be translocated from the extracellular space into the cytosol and nucleus of target cells, and regulates processes such as rRNA synthesis and cell growth. FGFR1 translocation requires p38 MAPK activation. In the nucleus, many transcription factors are phosphorylated and activated by p38 MAPKs in response to different stimuli. Classical examples include ATF1, ATF2, ATF6, ELK1, PTPRH, DDIT3, TP53/p53 and MEF2C and MEF2A. The p38 MAPKs are emerging as important modulators of gene expression by regulating chromatin modifiers and remodelers. The promoters of several genes involved in the inflammatory response, such as IL6, IL8 and IL12B, display a p38 MAPK-dependent enrichment of histone H3 phosphorylation on 'Ser-10' (H3S10ph) in LPS-stimulated myeloid cells. This phosphorylation enhances the accessibility of the cryptic NF-kappa-B-binding sites marking promoters for increased NF-kappa-B recruitment. Phosphorylates CDC25B and CDC25C which is required for binding to 14-3-3 proteins and leads to initiation of a G2 delay after ultraviolet radiation. Phosphorylates TIAR following DNA damage, releasing TIAR from GADD45A mRNA and preventing mRNA degradation. The p38 MAPKs may also have kinase-independent roles, which are thought to be due to the binding to targets in the absence of phosphorylation. Protein O-Glc-N-acylation catalyzed by the OGT is regulated by MAPK14, and, although OGT does not seem to be phosphorylated by MAPK14, their interaction increases upon MAPK14 activation induced by glucose deprivation. This interaction may regulate OGT activity by recruiting it to specific targets such as neurofilament H, stimulating its O-Glc-N-acylation. Required in mid-fetal development for the growth of embryo-derived blood vessels in the labyrinth layer of the placenta. Also plays an essential role in developmental and stress-induced erythropoiesis, through regulation of EPO gene expression. Isoform MXI2 activation is stimulated by mitogens and oxidative stress and only poorly phosphorylates ELK1 and ATF2. Isoform EXIP may play a role in the early onset of apoptosis. Phosphorylates S100A9 at 'Thr-113'. Serine/threonine kinase which acts as an essential component of the MAP kinase signal transduction pathway.
Click to Show/Hide
|
|||||
| BioChemical Class |
Kinase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.7.11.24
|
|||||
| Sequence |
MSQERPTFYRQELNKTIWEVPERYQNLSPVGSGAYGSVCAAFDTKTGLRVAVKKLSRPFQ
SIIHAKRTYRELRLLKHMKHENVIGLLDVFTPARSLEEFNDVYLVTHLMGADLNNIVKCQ KLTDDHVQFLIYQILRGLKYIHSADIIHRDLKPSNLAVNEDCELKILDFGLARHTDDEMT GYVATRWYRAPEIMLNWMHYNQTVDIWSVGCIMAELLTGRTLFPGTDHIDQLKLILRLVG TPGAELLKKISSESARNYIQSLTQMPKMNFANVFIGANPLAVDLLEKMLVLDSDKRITAA QALAHAYFAQYHDPDDEPVADPYDQSFESRDLLIDEWKSLTYDEVISFVPPPLDQEEMES Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| ADReCS ID | BADD_A00178 ; BADD_A05675 | |||||
| HIT2.0 ID | T71BDH | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Approved Drug(s) | [+] 1 Approved Drugs | + | ||||
| 1 | Ozagrel | Drug Info | Phase 4 | Xerophthalmia | [2] | |
| Clinical Trial Drug(s) | [+] 4 Clinical Trial Drugs | + | ||||
| 1 | Losmapimod | Drug Info | Phase 3 | Acute coronary syndrome | [3], [4] | |
| 2 | VX-702 | Drug Info | Phase 2a | Coronary artery disease | [5], [6] | |
| 3 | Dilmapimod | Drug Info | Phase 2 | Acute lung injury | [7], [8] | |
| 4 | VX-745 | Drug Info | Phase 2 | Rheumatoid arthritis | [9], [10], [11] | |
| Discontinued Drug(s) | [+] 7 Discontinued Drugs | + | ||||
| 1 | PAMAPIMOD | Drug Info | Discontinued in Phase 2 | Rheumatoid arthritis | [12] | |
| 2 | FR167653 | Drug Info | Discontinued in Phase 1 | Chronic obstructive pulmonary disease | [13] | |
| 3 | R-1487 | Drug Info | Discontinued in Phase 1 | Rheumatoid arthritis | [14] | |
| 4 | SB 235699 | Drug Info | Discontinued in Phase 1 | Psoriasis vulgaris | [15] | |
| 5 | SB-242235 | Drug Info | Discontinued in Phase 1 | Arthritis | [16] | |
| 6 | SB220025 | Drug Info | Terminated | Arthritis | [17], [18] | |
| 7 | SC-102 | Drug Info | Terminated | Solid tumour/cancer | [19] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Inhibitor | [+] 73 Inhibitor drugs | + | ||||
| 1 | Ozagrel | Drug Info | [1] | |||
| 2 | Losmapimod | Drug Info | [1] | |||
| 3 | Dilmapimod | Drug Info | [1] | |||
| 4 | VX-745 | Drug Info | [22], [23], [24] | |||
| 5 | PMID25991433-Compound-A1 | Drug Info | [25] | |||
| 6 | PMID25991433-Compound-F2 | Drug Info | [25] | |||
| 7 | PMID25991433-Compound-L1 | Drug Info | [25] | |||
| 8 | PMID25991433-Compound-L2 | Drug Info | [25] | |||
| 9 | PMID25991433-Compound-L3 | Drug Info | [25] | |||
| 10 | PMID25991433-Compound-O2 | Drug Info | [25] | |||
| 11 | PAMAPIMOD | Drug Info | [26] | |||
| 12 | FR167653 | Drug Info | [27], [28] | |||
| 13 | R-1487 | Drug Info | [29] | |||
| 14 | SB 235699 | Drug Info | [22], [30] | |||
| 15 | SB-242235 | Drug Info | [31] | |||
| 16 | SB 203580 | Drug Info | [32], [33] | |||
| 17 | SB220025 | Drug Info | [34] | |||
| 18 | SC-102 | Drug Info | [35] | |||
| 19 | (5-amino-1-phenyl-1H-pyrazol-4-yl)phenylmethanone | Drug Info | [36] | |||
| 20 | 2-Chlorophenol | Drug Info | [34] | |||
| 21 | 3-(1-NAPHTHYLMETHOXY)PYRIDIN-2-AMINE | Drug Info | [35] | |||
| 22 | 3-(Benzyloxy)Pyridin-2-Amine | Drug Info | [34] | |||
| 23 | 4,5,6,7-tetrabromo-1H-benzo[d][1,2,3]triazole | Drug Info | [37] | |||
| 24 | 4-(2-Ethyl-4-m-tolyl-thiazol-5-yl)-pyridine | Drug Info | [38] | |||
| 25 | 4-PHENOXY-N-(PYRIDIN-2-YLMETHYL)BENZAMIDE | Drug Info | [35] | |||
| 26 | 4-Phenylsulfanyl-7H-pyrrolo[2,3-d]pyrimidine | Drug Info | [39] | |||
| 27 | 4-[(3,5-diamino-1H-pyrazol-4-yl)diazenyl]phenol | Drug Info | [40] | |||
| 28 | 6-(4-Fluoro-phenylsulfanyl)-9H-purine | Drug Info | [39] | |||
| 29 | 6-Benzylsulfanyl-9H-purine | Drug Info | [39] | |||
| 30 | 6-o-tolylquinazolin-2-amine | Drug Info | [41] | |||
| 31 | 6-Phenylsulfanyl-9H-purine | Drug Info | [39] | |||
| 32 | 9-(4-Fluoro-benzyl)-6-phenylsulfanyl-9H-purine | Drug Info | [39] | |||
| 33 | 9-Benzyl-6-(4-fluoro-phenylsulfanyl)-9H-purine | Drug Info | [39] | |||
| 34 | 9-Benzyl-6-phenylsulfanyl-9H-purine | Drug Info | [39] | |||
| 35 | B-Octylglucoside | Drug Info | [34] | |||
| 36 | Bisindolylmaleimide-I | Drug Info | [42] | |||
| 37 | CI-1040 | Drug Info | [42] | |||
| 38 | Dihydro-quinolinone | Drug Info | [43] | |||
| 39 | DP-802 | Drug Info | [44] | |||
| 40 | GSK-280 | Drug Info | [44] | |||
| 41 | GW-788388 | Drug Info | [45] | |||
| 42 | IN-1130 | Drug Info | [46] | |||
| 43 | IN-1166 | Drug Info | [46] | |||
| 44 | KN-62 | Drug Info | [42] | |||
| 45 | KT-5720 | Drug Info | [42] | |||
| 46 | L-779450 | Drug Info | [47] | |||
| 47 | ML-3163 | Drug Info | [48] | |||
| 48 | ML-3375 | Drug Info | [49] | |||
| 49 | ML-3403 | Drug Info | [49] | |||
| 50 | N-(3-(trifluoromethoxy)benzyl)-4-phenoxybenzamide | Drug Info | [50] | |||
| 51 | N-(3-(trifluoromethyl)benzyl)-4-phenoxybenzamide | Drug Info | [50] | |||
| 52 | N-(4-(trifluoromethyl)benzyl)-4-phenoxybenzamide | Drug Info | [50] | |||
| 53 | N-(4-fluorobenzyl)-N-(pyridin-4-yl)-2-naphthamide | Drug Info | [51] | |||
| 54 | N-(4-methyl-benzyl)-4-phenoxy-benzamide | Drug Info | [50] | |||
| 55 | Oxindole 94 | Drug Info | [52] | |||
| 56 | PD-0166326 | Drug Info | [53] | |||
| 57 | PD-0173956 | Drug Info | [53] | |||
| 58 | PHA-666859 | Drug Info | [44] | |||
| 59 | Phenyl-(3-phenyl-1H-indazol-6-yl)-amine | Drug Info | [54] | |||
| 60 | PMID22521646C12 | Drug Info | [55] | |||
| 61 | RO-316233 | Drug Info | [42] | |||
| 62 | Ro-3201195 | Drug Info | [56] | |||
| 63 | Ro31-8220 | Drug Info | [42] | |||
| 64 | RWJ-68354 | Drug Info | [57] | |||
| 65 | SB-216995 | Drug Info | [43] | |||
| 66 | SB-218655 | Drug Info | [43] | |||
| 67 | SB-227931 | Drug Info | [58] | |||
| 68 | Small molecule 34 | Drug Info | [34] | |||
| 69 | STAUROSPORINONE | Drug Info | [42] | |||
| 70 | Triazolopyridine | Drug Info | [35] | |||
| 71 | Tyrphostin ag-1478 | Drug Info | [61] | |||
| 72 | UCB-1277763 | Drug Info | [44] | |||
| 73 | ZM-336372 | Drug Info | [62] | |||
| Modulator | [+] 2 Modulator drugs | + | ||||
| 1 | VX-702 | Drug Info | [20], [21] | |||
| 2 | Talmapimod | Drug Info | [59], [60] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Dasatinib | Ligand Info | |||||
| Structure Description | Human p38 MAP Kinase in Complex with Dasatinib | PDB:3LFA | ||||
| Method | X-ray diffraction | Resolution | 2.10 Å | Mutation | No | [63] |
| PDB Sequence |
ERPTFYRQEL
13 NKTIWEVPER23 YQNLSPVGSY35 GSVCAAFDTK45 TGLRVAVKKL55 SRPFQSIIHA 65 KRTYRELRLL75 KHMKHENVIG85 LLDVFTPARS95 LEEFNDVYLV105 THLMGADLNN 115 IVKCQKLTDD125 HVQFLIYQIL135 RGLKYIHSAD145 IIHRDLKPSN155 LAVNEDCELK 165 ILDATRWYRA190 PEIMLNWMHY200 NQTVDIWSVG210 CIMAELLTGR220 TLFPGTDHID 230 QLKLILRLVG240 TPGAELLKKI250 SSESARNYIQ260 SLTQMPKMNF270 ANVFIGANPL 280 AVDLLEKMLV290 LDSDKRITAA300 QALAHAYFAQ310 YHDPDDEPVA320 DPYDQSFESR 330 DLLIDEWKSL340 TYDEVISFVP350 PPL
|
|||||
|
|
||||||
| Ligand Name: Sorafenib | Ligand Info | |||||
| Structure Description | Human P38 MAP kinase in complex with Sorafenib | PDB:3GCS | ||||
| Method | X-ray diffraction | Resolution | 2.10 Å | Mutation | Yes | [64] |
| PDB Sequence |
RPTFYRQELN
14 KTIWEVPERY24 QNLSPVGYGS37 VCAAFDTKTG47 LRVAVKKLSR57 PFQSIIHAKR 67 TYRELRLLKH77 MKHENVIGLL87 DVFTPARSLE97 EFNDVYLVTH107 LMGADLNNIV 117 KSQKLTDDHV127 QFLIYQILRG137 LKYIHSADII147 HRDLKPSNLA157 VNEDSELKIL 167 DFGTRWYRAP191 EIMLNWMHYN201 QTVDIWSVGC211 IMAELLTGRT221 LFPGTDHIDQ 231 LKLILRLVGT241 PGAELLKKIS251 SESARNYIQS261 LTQMPKMNFA271 NVFIGANPLA 281 VDLLEKMLVL291 DSDKRITAAQ301 ALAHAYFAQY311 HDPDDEPVAD321 PYDQSLESRD 331 LLIDEWKSLT341 YDEVISFVPP351 P
|
|||||
|
|
VAL30
4.006
VAL38
3.625
ALA51
3.414
LYS53
4.202
ARG70
3.641
GLU71
2.561
LEU74
3.685
LEU75
3.880
MET78
3.955
VAL83
3.505
ILE84
3.318
LEU104
4.199
THR106
3.613
|
|||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| MAPK signaling pathway | hsa04010 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Rap1 signaling pathway | hsa04015 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| FoxO signaling pathway | hsa04068 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Sphingolipid signaling pathway | hsa04071 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Oocyte meiosis | hsa04114 | Affiliated Target |

|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Cellular senescence | hsa04218 | Affiliated Target |

|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Adrenergic signaling in cardiomyocytes | hsa04261 | Affiliated Target |

|
| Class: Organismal Systems => Circulatory system | Pathway Hierarchy | ||
| VEGF signaling pathway | hsa04370 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Osteoclast differentiation | hsa04380 | Affiliated Target |

|
| Class: Organismal Systems => Development and regeneration | Pathway Hierarchy | ||
| Signaling pathways regulating pluripotency of stem cells | hsa04550 | Affiliated Target |

|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
| Platelet activation | hsa04611 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Neutrophil extracellular trap formation | hsa04613 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Toll-like receptor signaling pathway | hsa04620 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| NOD-like receptor signaling pathway | hsa04621 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| RIG-I-like receptor signaling pathway | hsa04622 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| C-type lectin receptor signaling pathway | hsa04625 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
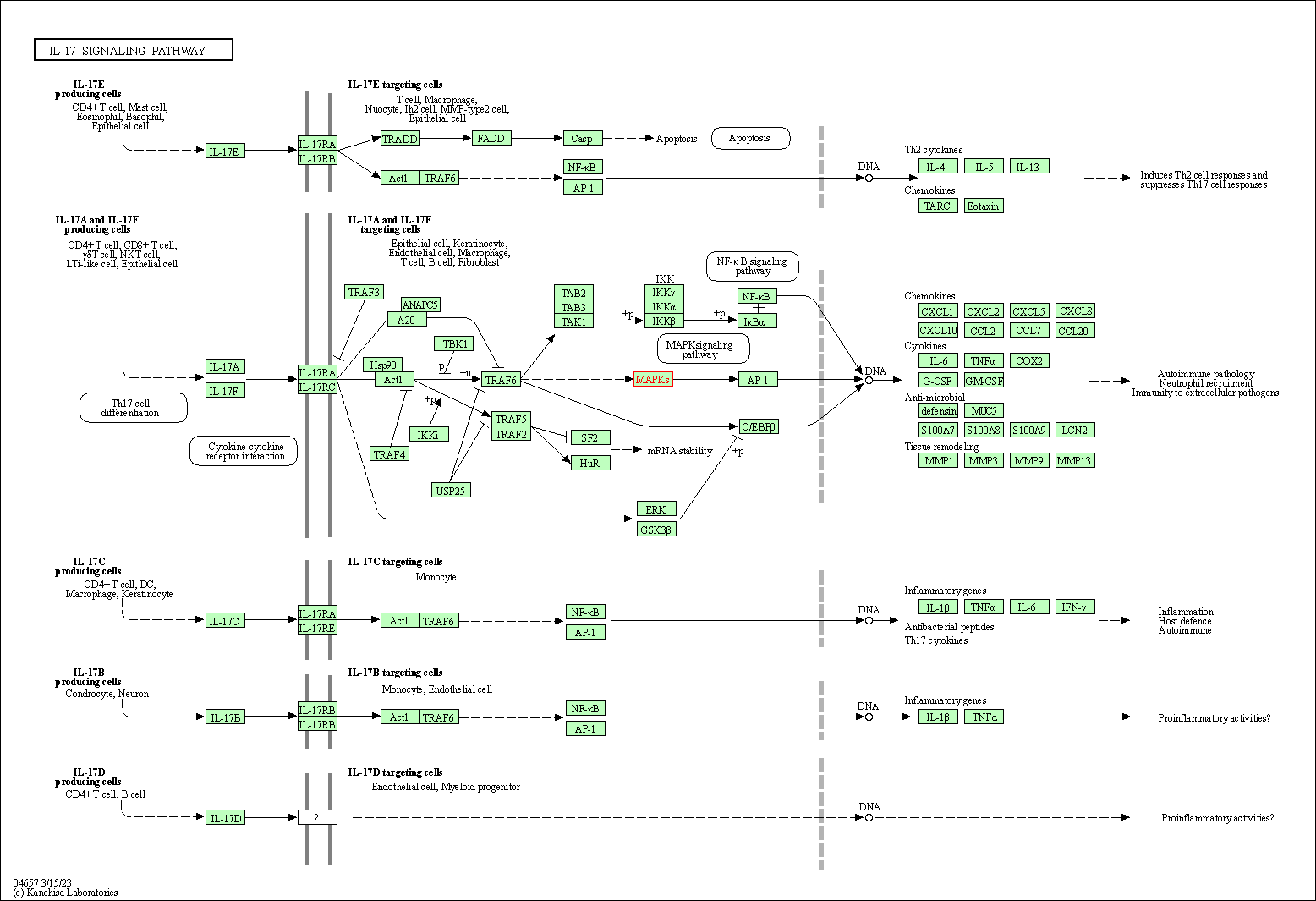
| IL-17 signaling pathway | hsa04657 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
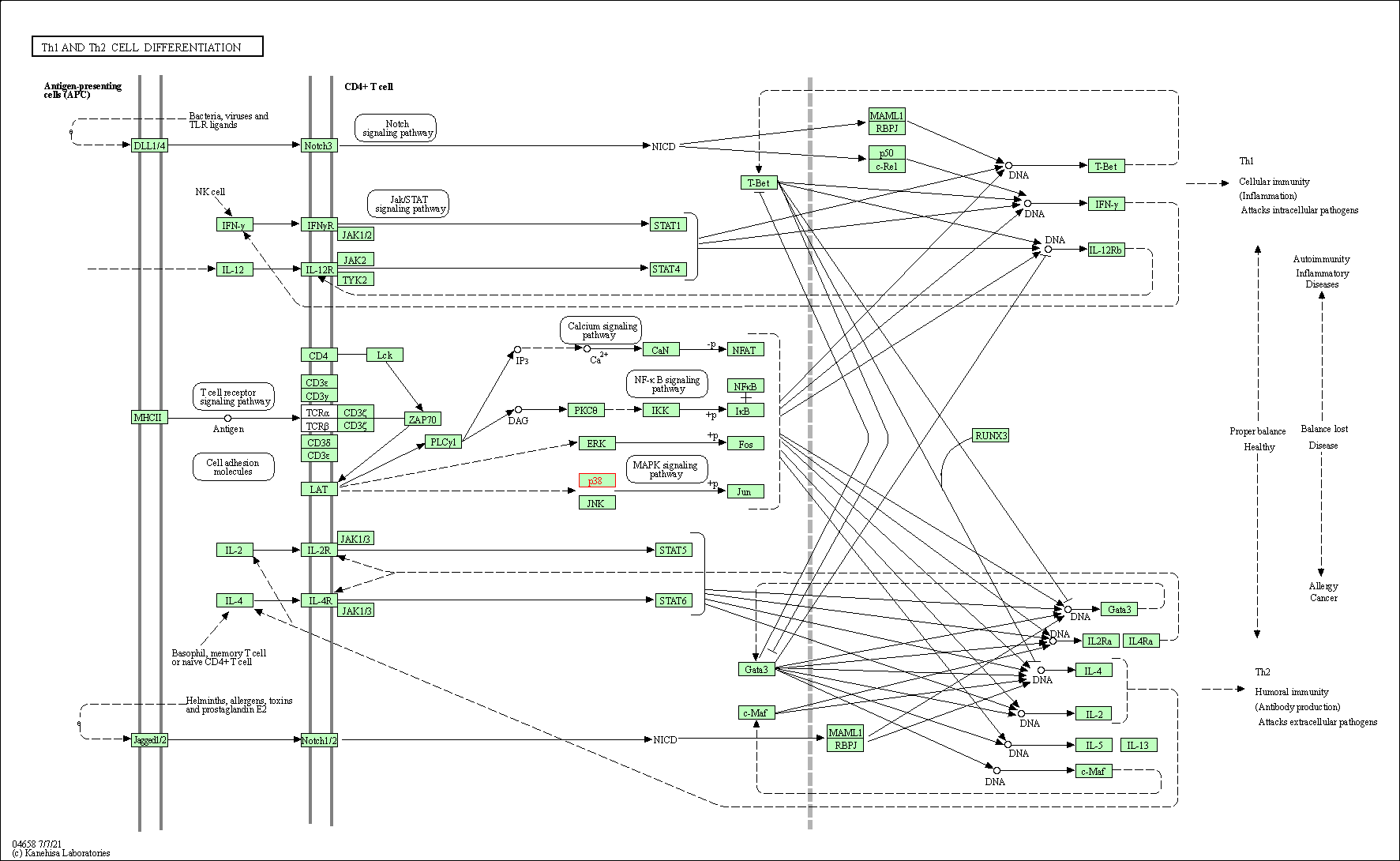
| Th1 and Th2 cell differentiation | hsa04658 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
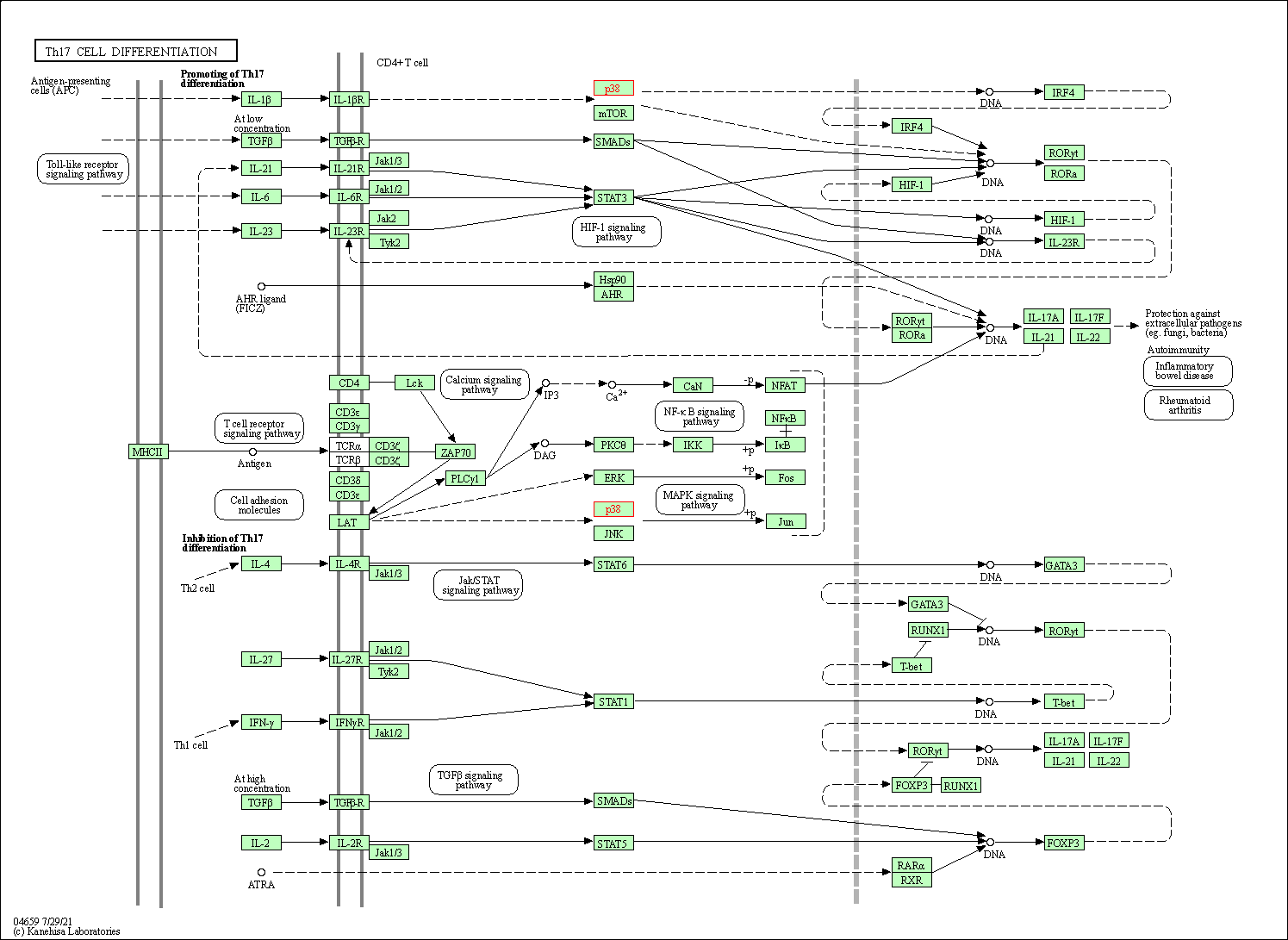
| Th17 cell differentiation | hsa04659 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
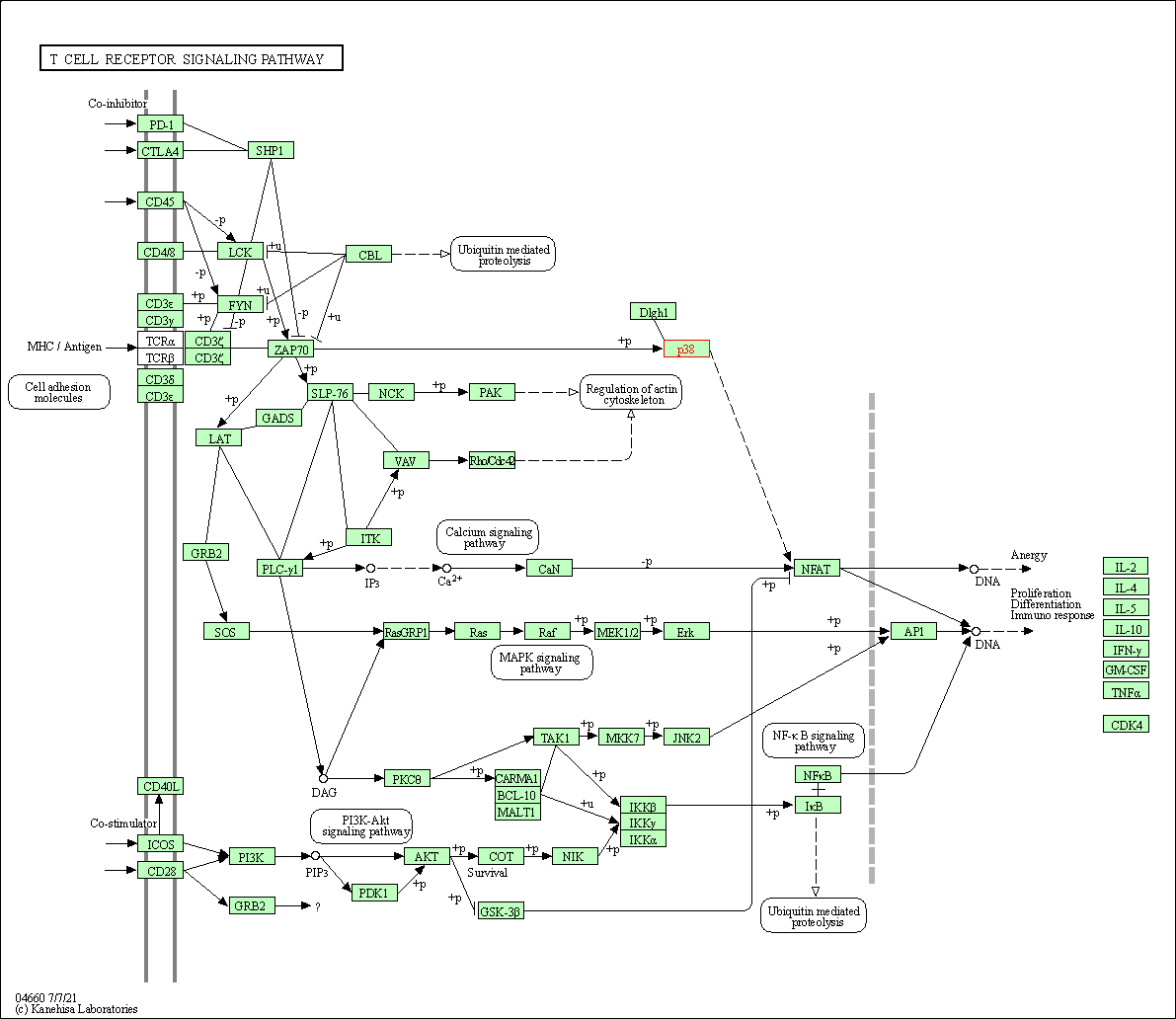
| T cell receptor signaling pathway | hsa04660 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
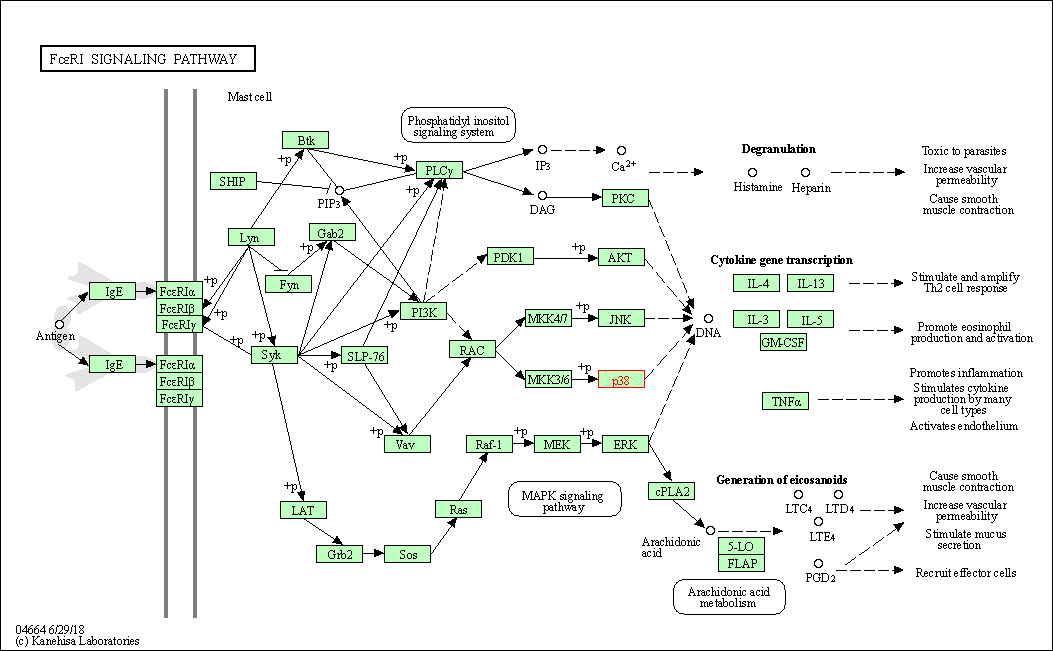
| Fc epsilon RI signaling pathway | hsa04664 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
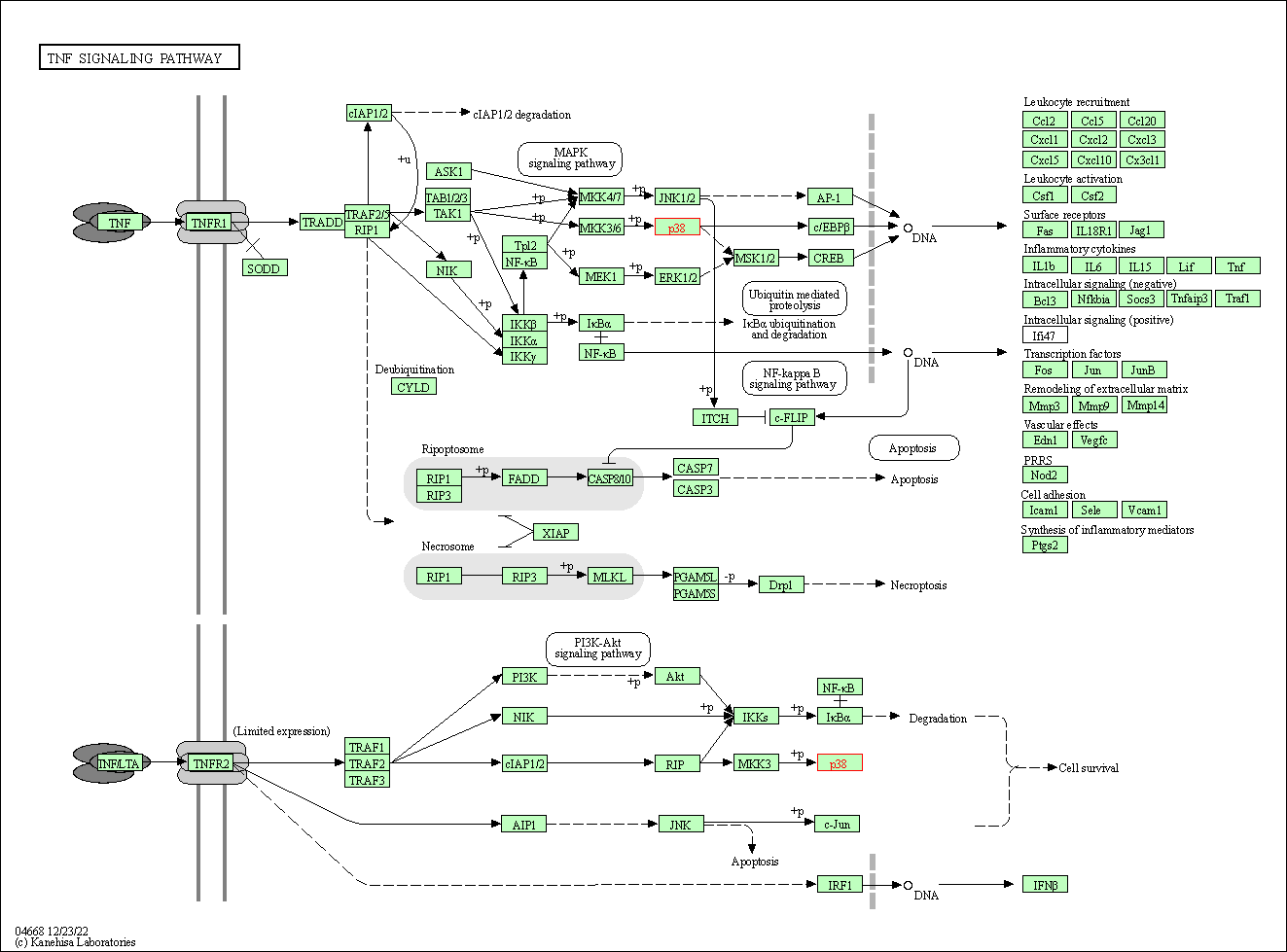
| TNF signaling pathway | hsa04668 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
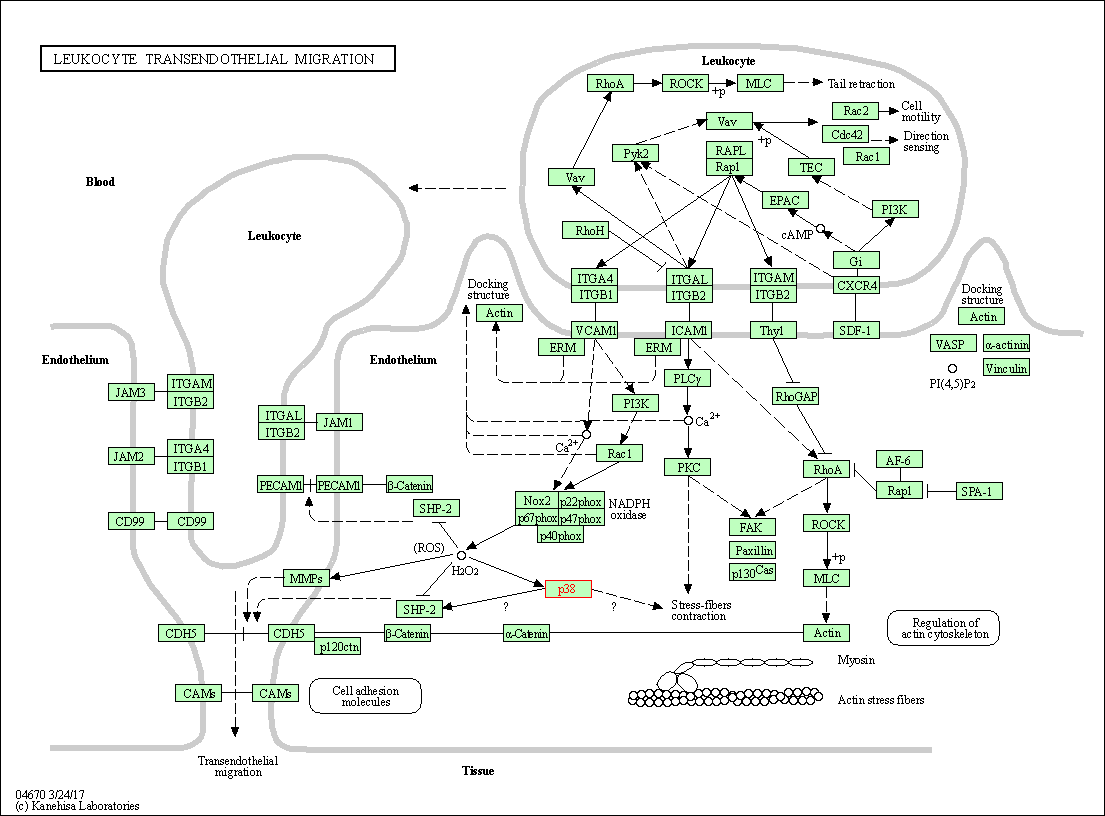
| Leukocyte transendothelial migration | hsa04670 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
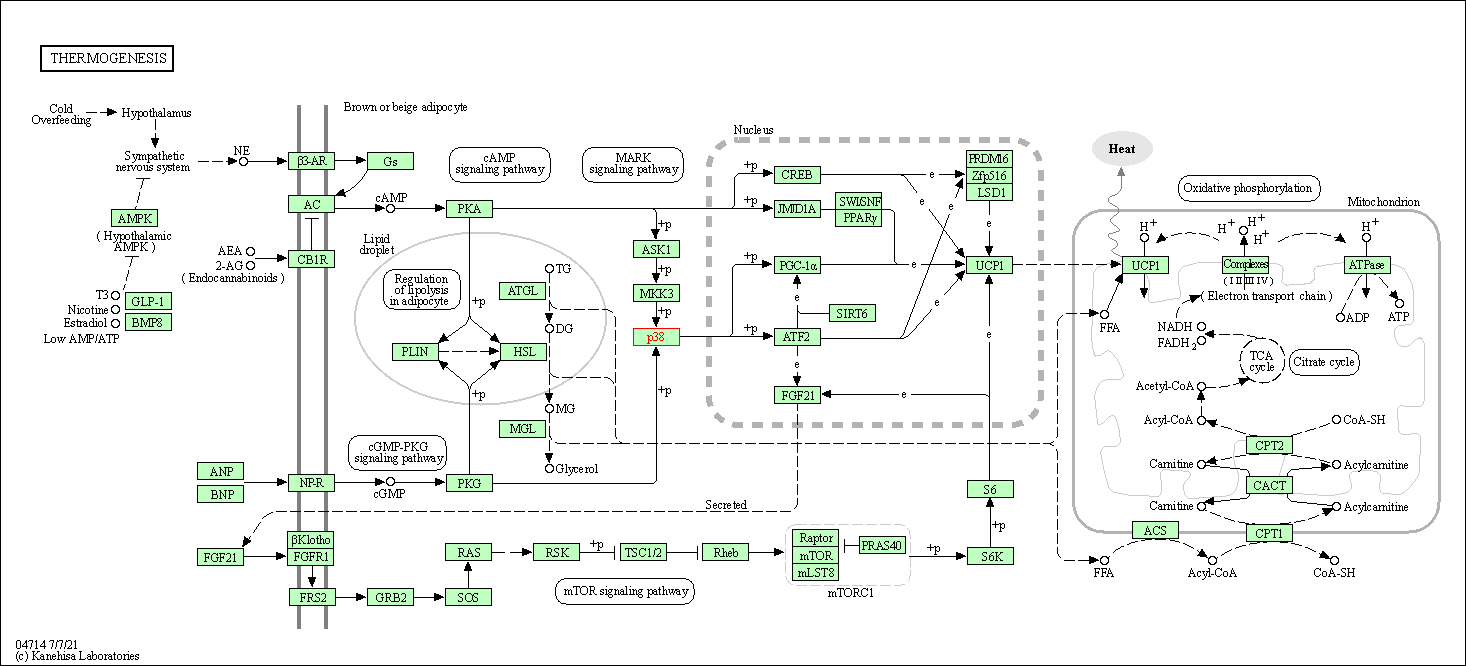
| Thermogenesis | hsa04714 | Affiliated Target |

|
| Class: Organismal Systems => Environmental adaptation | Pathway Hierarchy | ||
| Neurotrophin signaling pathway | hsa04722 | Affiliated Target |

|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Retrograde endocannabinoid signaling | hsa04723 | Affiliated Target |

|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Dopaminergic synapse | hsa04728 | Affiliated Target |

|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Inflammatory mediator regulation of TRP channels | hsa04750 | Affiliated Target |

|
| Class: Organismal Systems => Sensory system | Pathway Hierarchy | ||
| GnRH signaling pathway | hsa04912 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Progesterone-mediated oocyte maturation | hsa04914 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Prolactin signaling pathway | hsa04917 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Relaxin signaling pathway | hsa04926 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Growth hormone synthesis, secretion and action | hsa04935 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 61 | Degree centrality | 6.55E-03 | Betweenness centrality | 3.90E-03 |
|---|---|---|---|---|---|
| Closeness centrality | 2.67E-01 | Radiality | 1.46E+01 | Clustering coefficient | 9.78E-02 |
| Neighborhood connectivity | 4.28E+01 | Topological coefficient | 3.62E-02 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) |
||||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Clinical pipeline report, company report or official report of GlaxoSmithKline (2009). | |||||
| REF 2 | ClinicalTrials.gov (NCT00200356) Edaravone-Sodium Ozagrel Comparative Post-Marketing Study on Acute Ischemic Stroke. U.S. National Institutes of Health. | |||||
| REF 3 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7835). | |||||
| REF 4 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800023191) | |||||
| REF 5 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 6059). | |||||
| REF 6 | Emerging drugs for rheumatoid arthritis. Expert Opin Emerg Drugs. 2008 Mar;13(1):175-96. | |||||
| REF 7 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7815). | |||||
| REF 8 | ClinicalTrials.gov (NCT00291902) A Pharmacokinetic Study Of SB-681323 In Subjects With Coronary Heart Disease Undergoing Percutaneous Intervention. U.S. National Institutes of Health. | |||||
| REF 9 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 5719). | |||||
| REF 10 | ClinicalTrials.gov (NCT02423200) Clinical Pharmacology of p38 MAP Kinase Inhibitor, VX-745, in Mild Cognitive Impairment Due to Alzheimer's Disease (AD) or Mild AD. U.S. National Institutes of Health. | |||||
| REF 11 | Myelodysplastic Syndromes: Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2011 January; 9(1): 30-56. | |||||
| REF 12 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800018538) | |||||
| REF 13 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800012873) | |||||
| REF 14 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800017105) | |||||
| REF 15 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800012315) | |||||
| REF 16 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800010787) | |||||
| REF 17 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 6038). | |||||
| REF 18 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800010505) | |||||
| REF 19 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800016901) | |||||
| REF 20 | Efficacy, pharmacodynamics, and safety of VX-702, a novel p38 MAPK inhibitor, in rheumatoid arthritis: results of two randomized, double-blind, placebo-controlled clinical studies. Arthritis Rheum. 2009 May;60(5):1232-41. | |||||
| REF 21 | Comparative renal excretion of VX-702, a novel p38 MAPK inhibitor, and methotrexate in the perfused rat kidney model. Drug Dev Ind Pharm. 2010 Mar;36(3):315-22. | |||||
| REF 22 | Pharmacological inhibitors of MAPK pathways. Trends Pharmacol Sci. 2002 Jan;23(1):40-5. | |||||
| REF 23 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 24 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 25 | c-Jun N-terminal kinase inhibitors: a patent review (2010 - 2014).Expert Opin Ther Pat. 2015;25(8):849-72. | |||||
| REF 26 | Selective p38alpha inhibitors clinically evaluated for the treatment of chronic inflammatory disorders. J Med Chem. 2010 Mar 25;53(6):2345-53. | |||||
| REF 27 | Prevention of the onset and progression of collagen-induced arthritis in rats by the potent p38 mitogen-activated protein kinase inhibitor FR167653. Arthritis Rheum. 2003 Sep;48(9):2670-81. | |||||
| REF 28 | Involvement of p38 mitogen-activated protein kinase followed by chemokine expression in crescentic glomerulonephritis. Am J Kidney Dis. 2001 Dec;38(6):1169-77. | |||||
| REF 29 | Discovery of 6-(2,4-difluorophenoxy)-2-[3-hydroxy-1-(2-hydroxyethyl)propylamino]-8-methyl-8H-pyrido[2,3-d]pyrimidin-7-one (pamapimod) and 6-(2,4-difluorophenoxy)-8-methyl-2-(tetrahydro-2H-pyran-4-ylamino)pyrido[2,3-d]pyrimidin-7(8H)-one (R1487) as orally bioavailable and highly selective inhibitors of p38alpha mitogen-activated protein kinase. J Med Chem. 2011 Apr 14;54(7):2255-65. | |||||
| REF 30 | Synthesis and structure-activity relationship of aminobenzophenones. A novel class of p38 MAP kinase inhibitors with high antiinflammatory activity. J Med Chem. 2003 Dec 18;46(26):5651-62. | |||||
| REF 31 | Biphenyl amide p38 kinase inhibitors 4: DFG-in and DFG-out binding modes. Bioorg Med Chem Lett. 2008 Aug 1;18(15):4433-7. | |||||
| REF 32 | The p38 mitogen-activated protein kinase pathway plays a critical role in thrombin-induced endothelial chemokine production and leukocyte recruitment. Blood. 2001 Aug 1;98(3):667-73. | |||||
| REF 33 | Privileged structures: a useful concept for the rational design of new lead drug candidates. Mini Rev Med Chem. 2007 Nov;7(11):1108-19. | |||||
| REF 34 | How many drug targets are there Nat Rev Drug Discov. 2006 Dec;5(12):993-6. | |||||
| REF 35 | The Protein Data Bank. Nucleic Acids Res. 2000 Jan 1;28(1):235-42. | |||||
| REF 36 | Discovery of S-[5-amino-1-(4-fluorophenyl)-1H-pyrazol-4-yl]-[3-(2,3-dihydroxypropoxy)phenyl]methanone (RO3201195), an orally bioavailable and highl... J Med Chem. 2006 Mar 9;49(5):1562-75. | |||||
| REF 37 | Optimization of protein kinase CK2 inhibitors derived from 4,5,6,7-tetrabromobenzimidazole. J Med Chem. 2004 Dec 2;47(25):6239-47. | |||||
| REF 38 | Novel inhibitor of p38 MAP kinase as an anti-TNF-alpha drug: discovery of N-[4-[2-ethyl-4-(3-methylphenyl)-1,3-thiazol-5-yl]-2-pyridyl]benzamide (T... J Med Chem. 2005 Sep 22;48(19):5966-79. | |||||
| REF 39 | Synthesis and biological testing of purine derivatives as potential ATP-competitive kinase inhibitors. J Med Chem. 2005 Feb 10;48(3):710-22. | |||||
| REF 40 | 4-arylazo-3,5-diamino-1H-pyrazole CDK inhibitors: SAR study, crystal structure in complex with CDK2, selectivity, and cellular effects. J Med Chem. 2006 Nov 2;49(22):6500-9. | |||||
| REF 41 | Discovery of aminoquinazolines as potent, orally bioavailable inhibitors of Lck: synthesis, SAR, and in vivo anti-inflammatory activity. J Med Chem. 2006 Sep 21;49(19):5671-86. | |||||
| REF 42 | Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem J. 2000 Oct 1;351(Pt 1):95-105. | |||||
| REF 43 | In silico search for multi-target anti-inflammatories in Chinese herbs and formulas. Bioorg Med Chem. 2010 Mar 15;18(6):2204-2218. | |||||
| REF 44 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Target id: 1499). | |||||
| REF 45 | Discovery of 4-{4-[3-(pyridin-2-yl)-1H-pyrazol-4-yl]pyridin-2-yl}-N-(tetrahydro-2H- pyran-4-yl)benzamide (GW788388): a potent, selective, and orall... J Med Chem. 2006 Apr 6;49(7):2210-21. | |||||
| REF 46 | Synthesis and biological evaluation of trisubstituted imidazole derivatives as inhibitors of p38alpha mitogen-activated protein kinase. Bioorg Med Chem Lett. 2008 Jul 15;18(14):4006-10. | |||||
| REF 47 | The identification of potent and selective imidazole-based inhibitors of B-Raf kinase. Bioorg Med Chem Lett. 2006 Jan 15;16(2):378-81. | |||||
| REF 48 | From imidazoles to pyrimidines: new inhibitors of cytokine release. J Med Chem. 2002 Jun 20;45(13):2733-40. | |||||
| REF 49 | Novel substituted pyridinyl imidazoles as potent anticytokine agents with low activity against hepatic cytochrome P450 enzymes. J Med Chem. 2003 Jul 17;46(15):3230-44. | |||||
| REF 50 | Two classes of p38alpha MAP kinase inhibitors having a common diphenylether core but exhibiting divergent binding modes. Bioorg Med Chem Lett. 2005 Dec 1;15(23):5274-9. | |||||
| REF 51 | Amide-based inhibitors of p38alpha MAP kinase. Part 2: design, synthesis and SAR of potent N-pyrimidyl amides. Bioorg Med Chem Lett. 2010 Apr 15;20(8):2560-3. | |||||
| REF 52 | Inhibition of p38 MAPK decreases myocardial TNF-alpha expression and improves myocardial function and survival in endotoxemia. Cardiovasc Res. 2003 Oct 1;59(4):893-900. | |||||
| REF 53 | Structure-activity relationships of 6-(2,6-dichlorophenyl)-8-methyl-2-(phenylamino)pyrido[2,3-d]pyrimidin-7-ones: toward selective Abl inhibitors. Bioorg Med Chem Lett. 2009 Dec 15;19(24):6872-6. | |||||
| REF 54 | Design and synthesis of 6-anilinoindazoles as selective inhibitors of c-Jun N-terminal kinase-3. Bioorg Med Chem Lett. 2005 Nov 15;15(22):5095-9. | |||||
| REF 55 | Novel triazolopyridylbenzamides as potent and selective p38alpha inhibitors. Bioorg Med Chem Lett. 2012 May 15;22(10):3431-6. | |||||
| REF 56 | Microwave-assisted synthesis of 5-aminopyrazol-4-yl ketones and the p38(MAPK) inhibitor RO3201195 for study in Werner syndrome cells. Bioorg Med Chem Lett. 2008 Jul 1;18(13):3745-8. | |||||
| REF 57 | Imidazopyrimidines, potent inhibitors of p38 MAP kinase. Bioorg Med Chem Lett. 2003 Feb 10;13(3):347-50. | |||||
| REF 58 | The neuroprotective action of JNK3 inhibitors based on the 6,7-dihydro-5H-pyrrolo[1,2-a]imidazole scaffold. Bioorg Med Chem Lett. 2005 Nov 1;15(21):4666-70. | |||||
| REF 59 | Mitogen-activated protein kinases in innate immunity.Nat Rev Immunol.2013 Sep;13(9):679-92. | |||||
| REF 60 | p38 MAP Kinase Inhibition Reduces Propionibacterium acnes-Induced Inflammation in Vitro. Dermatol Ther (Heidelb). 2015 Mar;5(1):53-66. | |||||
| REF 61 | Novel, potent and selective anilinoquinazoline and anilinopyrimidine inhibitors of p38 MAP kinase. Bioorg Med Chem Lett. 2004 Nov 1;14(21):5389-94. | |||||
| REF 62 | A novel series of p38 MAP kinase inhibitors for the potential treatment of rheumatoid arthritis. Bioorg Med Chem Lett. 2004 Nov 1;14(21):5383-7. | |||||
| REF 63 | Development of novel thiazole-urea compounds which stabalize the inactive conformation of p38 alpha | |||||
| REF 64 | Development of a fluorescent-tagged kinase assay system for the detection and characterization of allosteric kinase inhibitors. J Am Chem Soc. 2009 Sep 23;131(37):13286-96. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

