Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T62390
(Former ID: TTDS00365)
|
|||||
| Target Name |
Tyrosine 3-monooxygenase (TH)
|
|||||
| Synonyms |
Tyrosine 3-hydroxylase; TH
Click to Show/Hide
|
|||||
| Gene Name |
TH
|
|||||
| Target Type |
Successful target
|
[1] | ||||
| Disease | [+] 2 Target-related Diseases | + | ||||
| 1 | Adrenomedullary hyperfunction [ICD-11: 5A75] | |||||
| 2 | Nutritional deficiency [ICD-11: 5B50-5B71] | |||||
| Function |
Plays an important role in the physiology of adrenergic neurones.
Click to Show/Hide
|
|||||
| BioChemical Class |
Paired donor oxygen oxidoreductase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 1.14.16.2
|
|||||
| Sequence |
MPTPDATTPQAKGFRRAVSELDAKQAEAIMVRGQGAPGPSLTGSPWPGTAAPAASYTPTP
RSPRFIGRRQSLIEDARKEREAAVAAAAAAVPSEPGDPLEAVAFEEKEGKAVLNLLFSPR ATKPSALSRAVKVFETFEAKIHHLETRPAQRPRAGGPHLEYFVRLEVRRGDLAALLSGVR QVSEDVRSPAGPKVPWFPRKVSELDKCHHLVTKFDPDLDLDHPGFSDQVYRQRRKLIAEI AFQYRHGDPIPRVEYTAEEIATWKEVYTTLKGLYATHACGEHLEAFALLERFSGYREDNI PQLEDVSRFLKERTGFQLRPVAGLLSARDFLASLAFRVFQCTQYIRHASSPMHSPEPDCC HELLGHVPMLADRTFAQFSQDIGLASLGASDEEIEKLSTLYWFTVEFGLCKQNGEVKAYG AGLLSSYGELLHCLSEEPEIRAFDPEAAAVQPYQDQTYQSVYFVSESFSDAKDKLRSYAS RIQRPFSVKFDPYTLAIDVLDSPQAVRRSLEGVQDELDTLAHALSAIG Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| HIT2.0 ID | T23TJV | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Approved Drug(s) | [+] 4 Approved Drugs | + | ||||
| 1 | L-Phenylalanine | Drug Info | Approved | Malnutrition | [2], [3] | |
| 2 | L-Tyrosine | Drug Info | Approved | Malnutrition | [4], [5] | |
| 3 | Metyrosine | Drug Info | Approved | Pheochromocytoma | [6], [7] | |
| 4 | Tetrahydrobiopterin | Drug Info | Approved | Malnutrition | [8], [9] | |
| Clinical Trial Drug(s) | [+] 3 Clinical Trial Drugs | + | ||||
| 1 | L1-79 | Drug Info | Phase 2 | Autism spectrum disorder | [10] | |
| 2 | OXB-102 | Drug Info | Phase 1/2 | Parkinson disease | [11] | |
| 3 | ProSavin | Drug Info | Phase 1/2 | Parkinson disease | [12] | |
| Mode of Action | [+] 4 Modes of Action | + | ||||
| Binder | [+] 3 Binder drugs | + | ||||
| 1 | L-Phenylalanine | Drug Info | [13], [14], [15] | |||
| 2 | L-Tyrosine | Drug Info | [14], [15] | |||
| 3 | Metyrosine | Drug Info | [1] | |||
| Cofactor | [+] 1 Cofactor drugs | + | ||||
| 1 | Tetrahydrobiopterin | Drug Info | [16] | |||
| Inhibitor | [+] 6 Inhibitor drugs | + | ||||
| 1 | L1-79 | Drug Info | [17] | |||
| 2 | 3-chlorotyrosine | Drug Info | [20] | |||
| 3 | 3-iodotyrosine | Drug Info | [20] | |||
| 4 | 7,8-dihydrobiopterin | Drug Info | [21] | |||
| 5 | alpha-propyldopacetamide | Drug Info | [20] | |||
| 6 | Meta-Tyrosine | Drug Info | [21] | |||
| Modulator | [+] 1 Modulator drugs | + | ||||
| 1 | ProSavin | Drug Info | [19] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Dopamine | Ligand Info | |||||
| Structure Description | Atomic model of the EM-based structure of the full-length tyrosine hydroxylase in complex with dopamine (residues 40-497) in which the regulatory domain (residues 40-165) has been included only with the backbone atoms | PDB:6ZVP | ||||
| Method | Electron microscopy | Resolution | 4.00 Å | Mutation | No | [22] |
| PDB Sequence |
SLIEDARKER
49 EAAVAAAAAA59 VPSEPGDPLE69 AVAFEEKEGK79 AMLNLLFSPR89 ATKPSALSRA 99 VKVFETFEAK109 IHHLETRPAQ119 RPRAGGPHLE129 YFVRLEVRRG139 DLAALLSGVR 149 QVSEDVRSPA159 GPKVPWFPRK169 VSELDKCHHL179 VTKFDPDLDL189 DHPGFSDQVY 199 RQRRKLIAEI209 AFQYRHGDPI219 PRVEYTAEEI229 ATWKEVYTTL239 KGLYATHACG 249 EHLEAFALLE259 RFSGYREDNI269 PQLEDVSRFL279 KERTGFQLRP289 VAGLLSARDF 299 LASLAFRVFQ309 CTQYIRHASS319 PMHSPEPDCC329 HELLGHVPML339 ADRTFAQFSQ 349 DIGLASLGAS359 DEEIEKLSTL369 YWFTVEFGLC379 KQNGEVKAYG389 AGLLSSYGEL 399 LHCLSEEPEI409 RAFDPEAAAV419 QPYQDQTYQS429 VYFVSESFSD439 AKDKLRSYAS 449 RIQRPFSVKF459 DPYTLAIDVL469 DSPQAVRRSL479 EGVQDELDTL489 AHALSAIG |
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
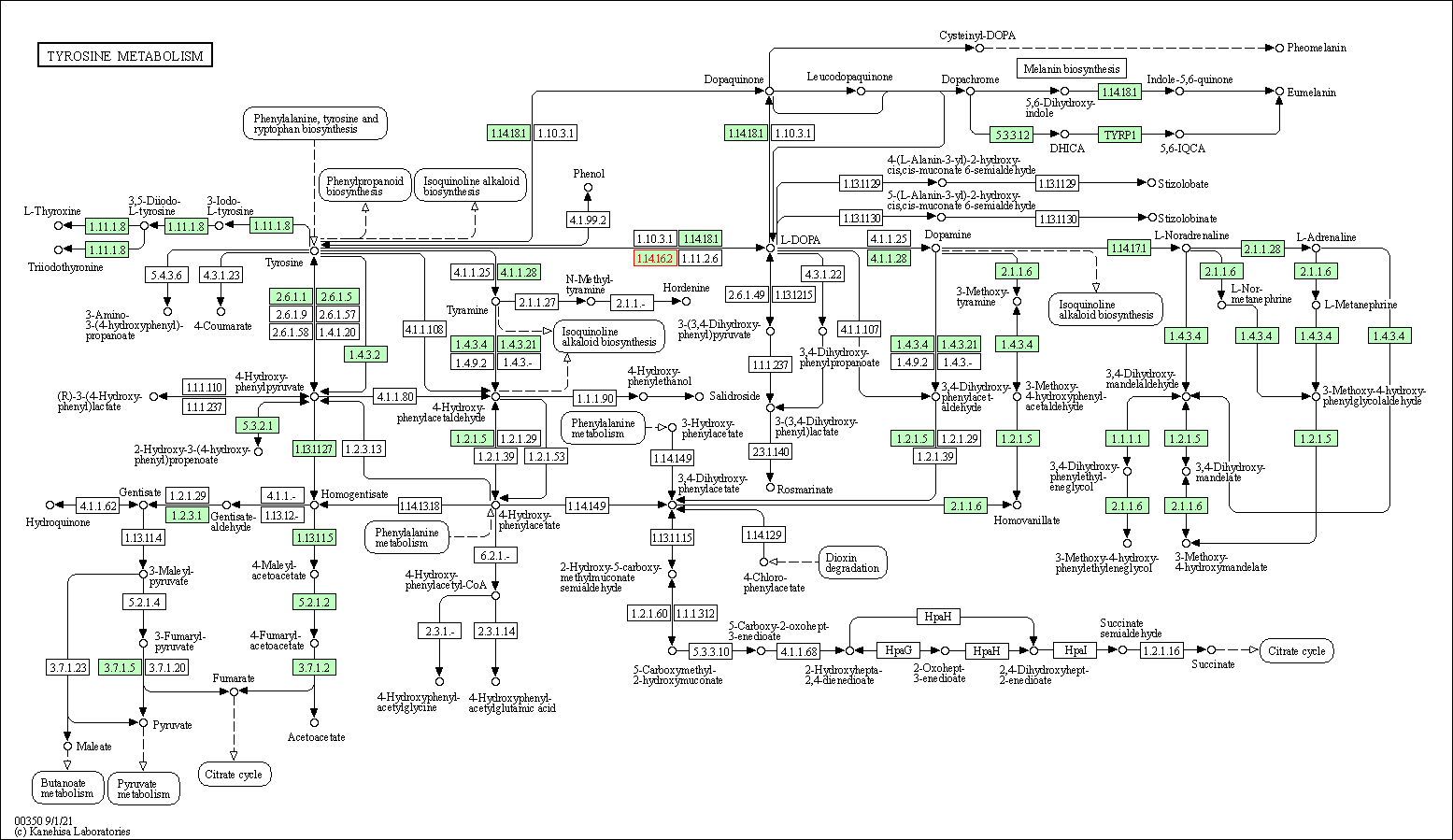
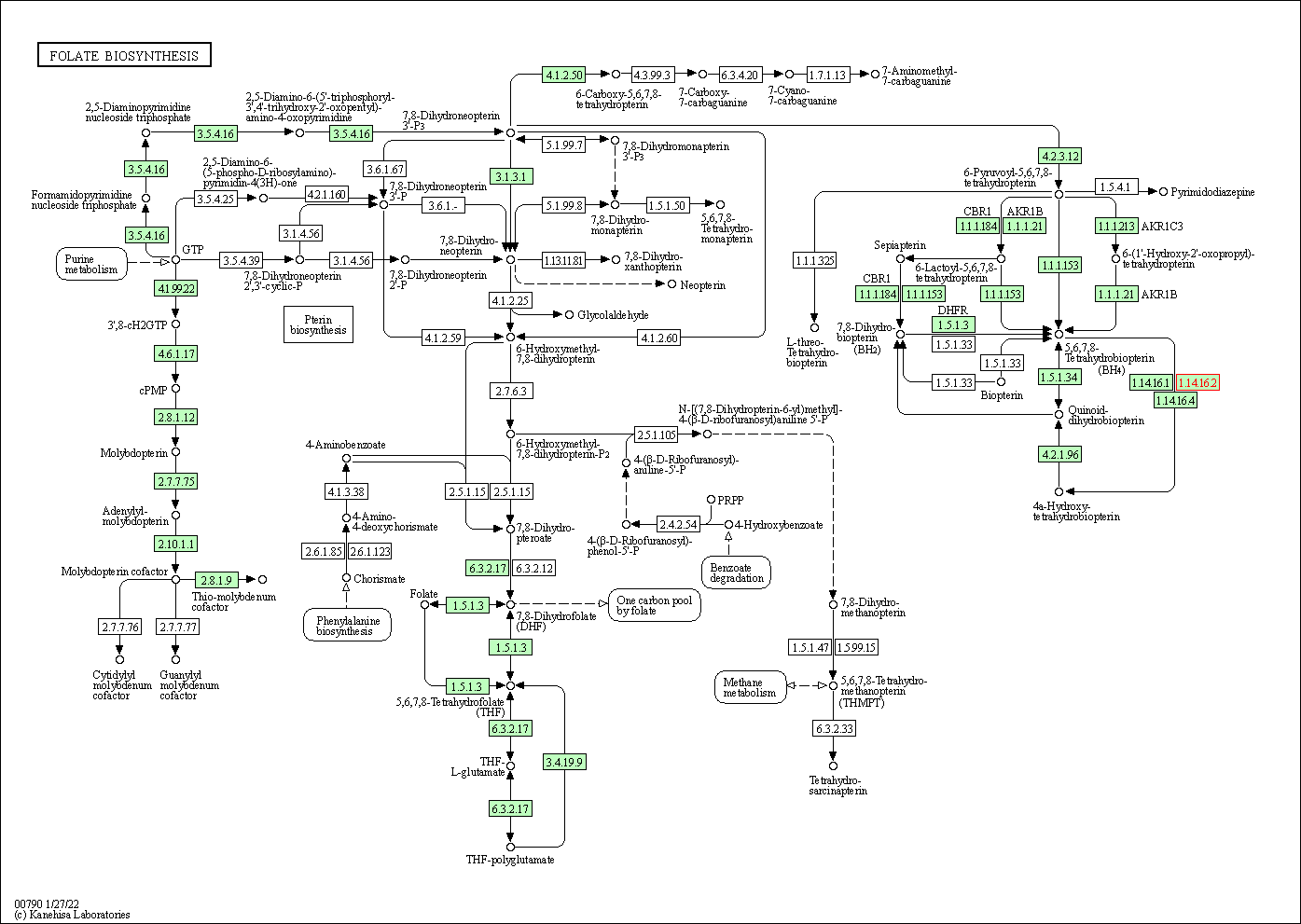
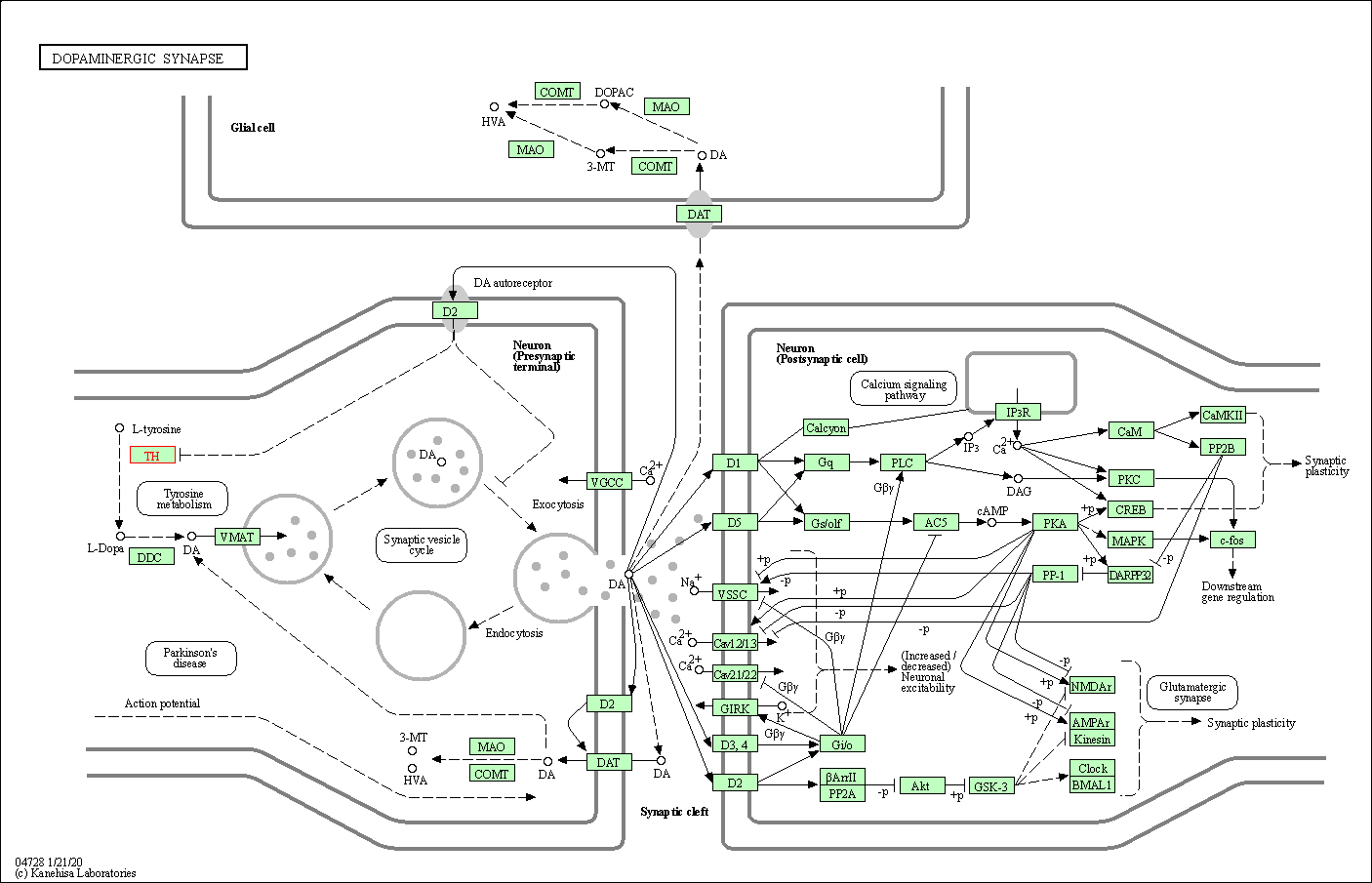
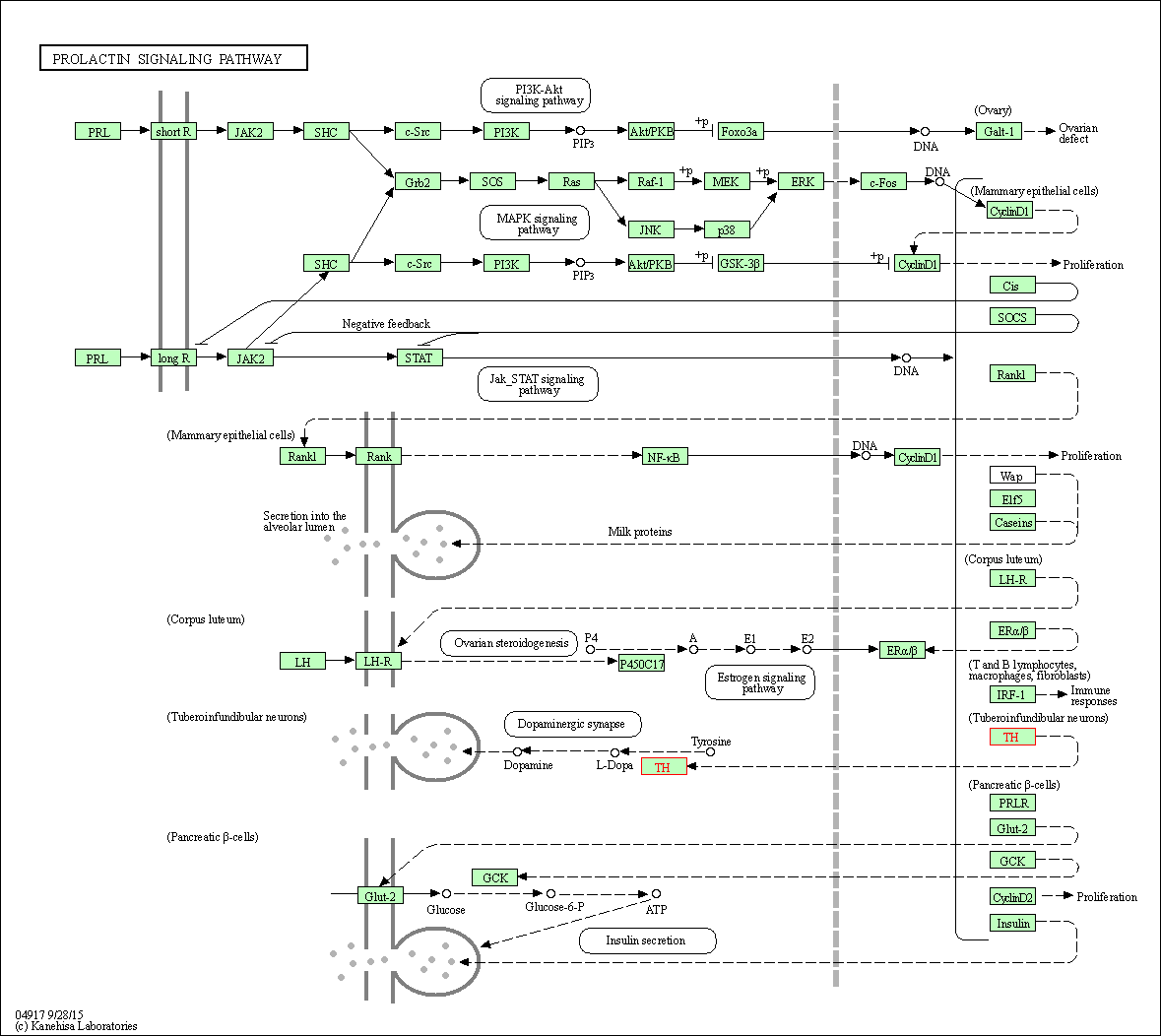
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Tyrosine metabolism | hsa00350 | Affiliated Target |

|
| Class: Metabolism => Amino acid metabolism | Pathway Hierarchy | ||
| Folate biosynthesis | hsa00790 | Affiliated Target |

|
| Class: Metabolism => Metabolism of cofactors and vitamins | Pathway Hierarchy | ||
| Dopaminergic synapse | hsa04728 | Affiliated Target |

|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Prolactin signaling pathway | hsa04917 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Degree | 5 | Degree centrality | 5.37E-04 | Betweenness centrality | 8.75E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 2.01E-01 | Radiality | 1.35E+01 | Clustering coefficient | 2.00E-01 |
| Neighborhood connectivity | 1.34E+01 | Topological coefficient | 2.41E-01 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) |
||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Dopamine beta-hydroxylase deficiency. A genetic disorder of cardiovascular regulation. Hypertension. 1991 Jul;18(1):1-8. | |||||
| REF 2 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 3313). | |||||
| REF 3 | Drug information of L-Alanine, Health Canada, 2008. (drugid: 1025) | |||||
| REF 4 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 4791). | |||||
| REF 5 | Drug information of L-Phenylalanine, 2008. eduDrugs. | |||||
| REF 6 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 6956). | |||||
| REF 7 | FDA Approved Drug Products from FDA Official Website. 2009. Application Number: (NDA) 017871. | |||||
| REF 8 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 5276). | |||||
| REF 9 | Sapropterin: a new therapeutic agent for phenylketonuria. Ann Pharmacother. 2009 Sep;43(9):1466-73. | |||||
| REF 10 | ClinicalTrials.gov (NCT02947048) Safety of L1-79 in Autism. U.S. National Institutes of Health. | |||||
| REF 11 | ClinicalTrials.gov (NCT03720418) Study of OXB-102 (AXO-Lenti-PD) in Patients With Bilateral, Idiopathic Parkinson's Disease (SUNRISE-PD). U.S. National Institutes of Health. | |||||
| REF 12 | ClinicalTrials.gov (NCT01856439) Long Term Safety and Efficacy Study of ProSavin in Parkinson's Disease. U.S. National Institutes of Health. | |||||
| REF 13 | Selectivity and affinity determinants for ligand binding to the aromatic amino acid hydroxylases. Curr Med Chem. 2007;14(4):455-67. | |||||
| REF 14 | Effect of metals and phenylalanine on the activity of human tryptophan hydroxylase-2: comparison with that on tyrosine hydroxylase activity. Neurosci Lett. 2006 Jul 3;401(3):261-5. | |||||
| REF 15 | In situ and in vitro evidence for DCoH/HNF-1 alpha transcription of tyrosinase in human skin melanocytes. Biochem Biophys Res Commun. 2003 Feb 7;301(2):610-6. | |||||
| REF 16 | Biochemistry of postmortem brains in Parkinson's disease: historical overview and future prospects. J Neural Transm Suppl. 2007;(72):113-20. | |||||
| REF 17 | Effect of L1-79 on Core Symptoms of Autism Spectrum Disorder: A Case Series. Clin Ther. 2019 Oct;41(10):1972-1981. | |||||
| REF 18 | Clinical pipeline report, company report or official report of Oxford BioMedica. | |||||
| REF 19 | Clinical pipeline report, company report or official report of Oxford BioMedica. | |||||
| REF 20 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Target id: 1243). | |||||
| REF 21 | How many drug targets are there Nat Rev Drug Discov. 2006 Dec;5(12):993-6. | |||||
| REF 22 | Structural mechanism for tyrosine hydroxylase inhibition by dopamine and reactivation by Ser40 phosphorylation. Nat Commun. 2022 Jan 10;13(1):74. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

