Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T59480
|
|||||
| Target Name |
KRAS G12C mutant (KRAS G12C)
|
|||||
| Gene Name |
KRAS
|
|||||
| Target Type |
Successful target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Non-small-cell lung cancer [ICD-11: 2C25] | |||||
| UniProt ID | ||||||
| Sequence |
MTEYKLVVVGAGGVGKSALTIQLIQNHFVDEYDPTIEDSYRKQVVIDGETCLLDILDTAG
QEEYSAMRDQYMRTGEGFLCVFAINNTKSFEDIHHYREQIKRVKDSEDVPMVLVGNKCDL PSRTVDTKQAQDLARSYGIPFIETSAKTRQRVEDAFYTLVREIRQYRLKKISKEEKTPGC VKIKKCIIM Click to Show/Hide
|
|||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Approved Drug(s) | [+] 2 Approved Drugs | + | ||||
| 1 | Adagrasib | Drug Info | Approved | Non-small-cell lung cancer | [2] | |
| 2 | Sotorasib | Drug Info | Approved | Non-small-cell lung cancer | [3] | |
| Clinical Trial Drug(s) | [+] 4 Clinical Trial Drugs | + | ||||
| 1 | JDQ443 | Drug Info | Phase 3 | Non-small-cell lung cancer | [4] | |
| 2 | LY3499446 | Drug Info | Phase 1/2 | Solid tumour/cancer | [5] | |
| 3 | JNJ-74699157 | Drug Info | Phase 1 | Solid tumour/cancer | [6] | |
| 4 | RG6330 | Drug Info | Phase 1 | Solid tumour/cancer | [7] | |
| Mode of Action | [+] 1 Modes of Action | + | ||||
| Inhibitor | [+] 6 Inhibitor drugs | + | ||||
| 1 | Adagrasib | Drug Info | [8] | |||
| 2 | Sotorasib | Drug Info | [1] | |||
| 3 | JDQ443 | Drug Info | [9] | |||
| 4 | LY3499446 | Drug Info | [10] | |||
| 5 | JNJ-74699157 | Drug Info | [10] | |||
| 6 | RG6330 | Drug Info | [7] | |||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Guanosine-5'-Diphosphate | Ligand Info | |||||
| Structure Description | CRYSTAL STRUCTURE OF KRAS-G12C IN COMPLEX WITH COMPOUND 23 (BI-0474) | PDB:8AFB | ||||
| Method | X-ray diffraction | Resolution | 1.12 Å | Mutation | Yes | [11] |
| PDB Sequence |
TEYKLVVVGA
11 CGVGKSALTI21 QLIQNHFVDE31 YDPTIEDSYR41 KQVVIDGETC51 LLDILDTAGQ 61 EEYSAMRDQY71 MRTGEGFLCV81 FAINNTKSFE91 DIHHYREQIK101 RVKDSEDVPM 111 VLVGNKSDLP121 SRTVDTKQAQ131 DLARSYGIPF141 IETSAKTRQG151 VDDAFYTLVR 161 EIRK
|
|||||
|
|
ALA11
3.830
CYS12
3.663
GLY13
2.764
VAL14
3.355
GLY15
3.006
LYS16
2.782
SER17
2.931
ALA18
2.887
PHE28
4.030
VAL29
4.969
ASP30
2.686
GLU31
4.674
|
|||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Ligand Name: K-Ras(G12C) inhibitor 6 | Ligand Info | |||||
| Structure Description | Crystal Structure of small molecule disulfide 6 bound to K-Ras G12C | PDB:4LUC | ||||
| Method | X-ray diffraction | Resolution | 1.29 Å | Mutation | Yes | [12] |
| PDB Sequence |
MTEYKLVVVG
10 ACGVGKSALT20 IQLIQNHFVD30 EYDPTIEDSY40 RKQVVIDGET50 SLLDILDTAG 60 QEEYSAMRDQ70 YMRTGEGFLL80 VFAINNTKSF90 EDIHHYREQI100 KRVKDSEDVP 110 MVLVGNKSDL120 PSRTVDTKQA130 QDLARSYGIP140 FIETSAKTRQ150 GVDDAFYTLV 160 REIRKHKE
|
|||||
|
|
VAL7
2.861
VAL9
2.719
GLY10
2.737
ALA11
2.567
CYS12
2.035
GLY13
3.393
LYS16
3.981
PRO34
3.242
THR35
4.735
LEU56
4.241
THR58
2.390
ALA59
4.071
|
|||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Human Similarity Proteins
Human Pathway Affiliation
|
|
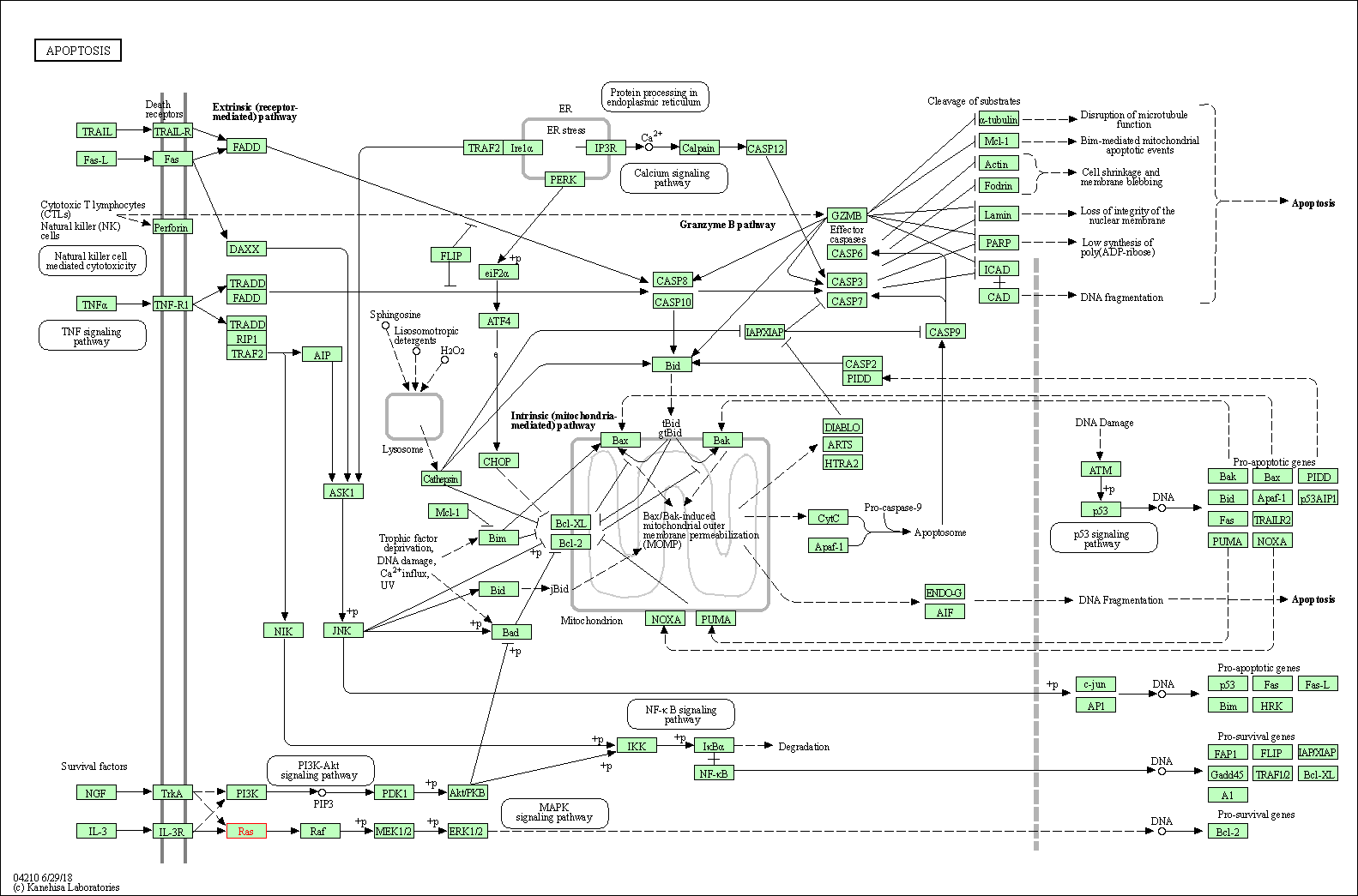
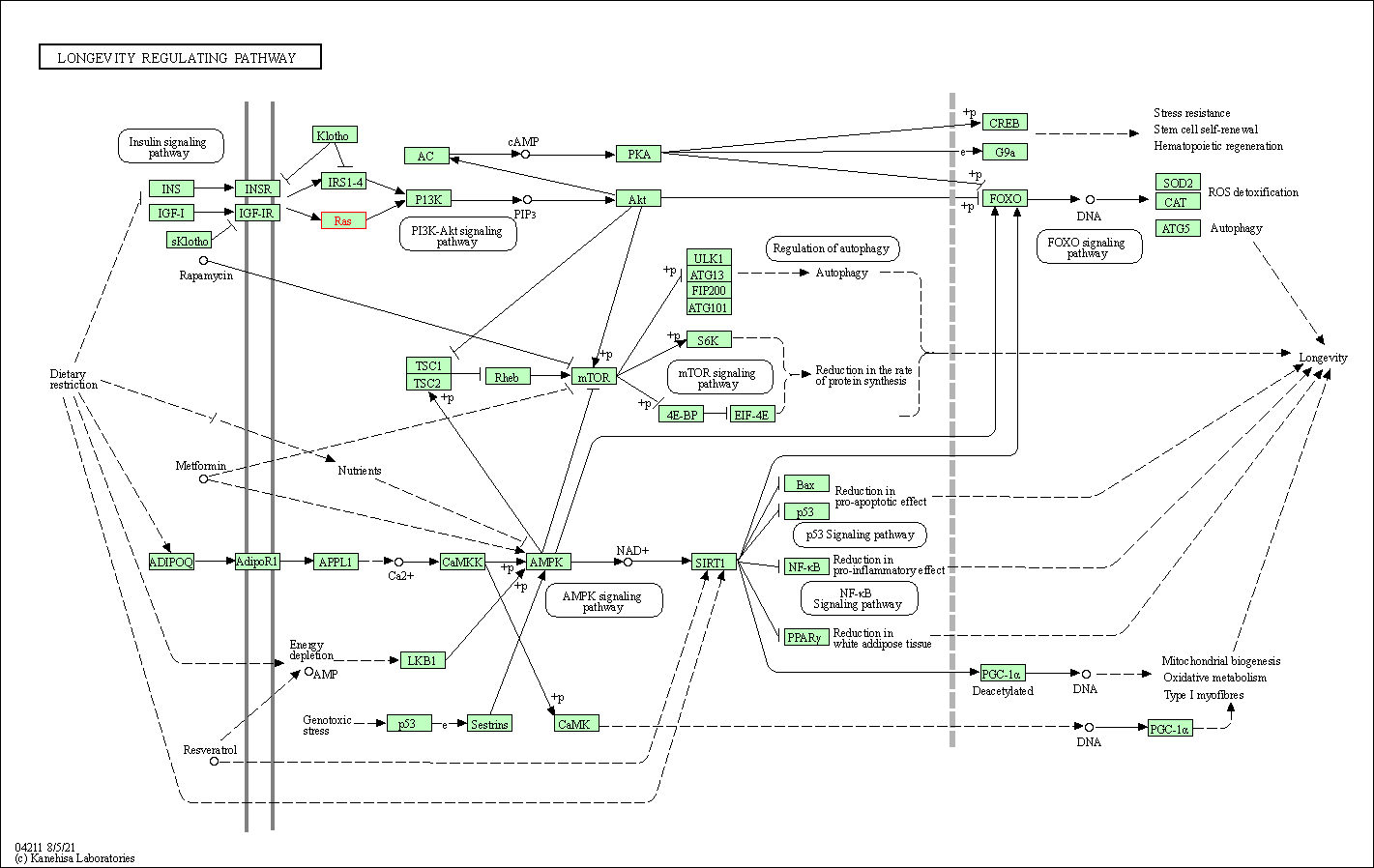
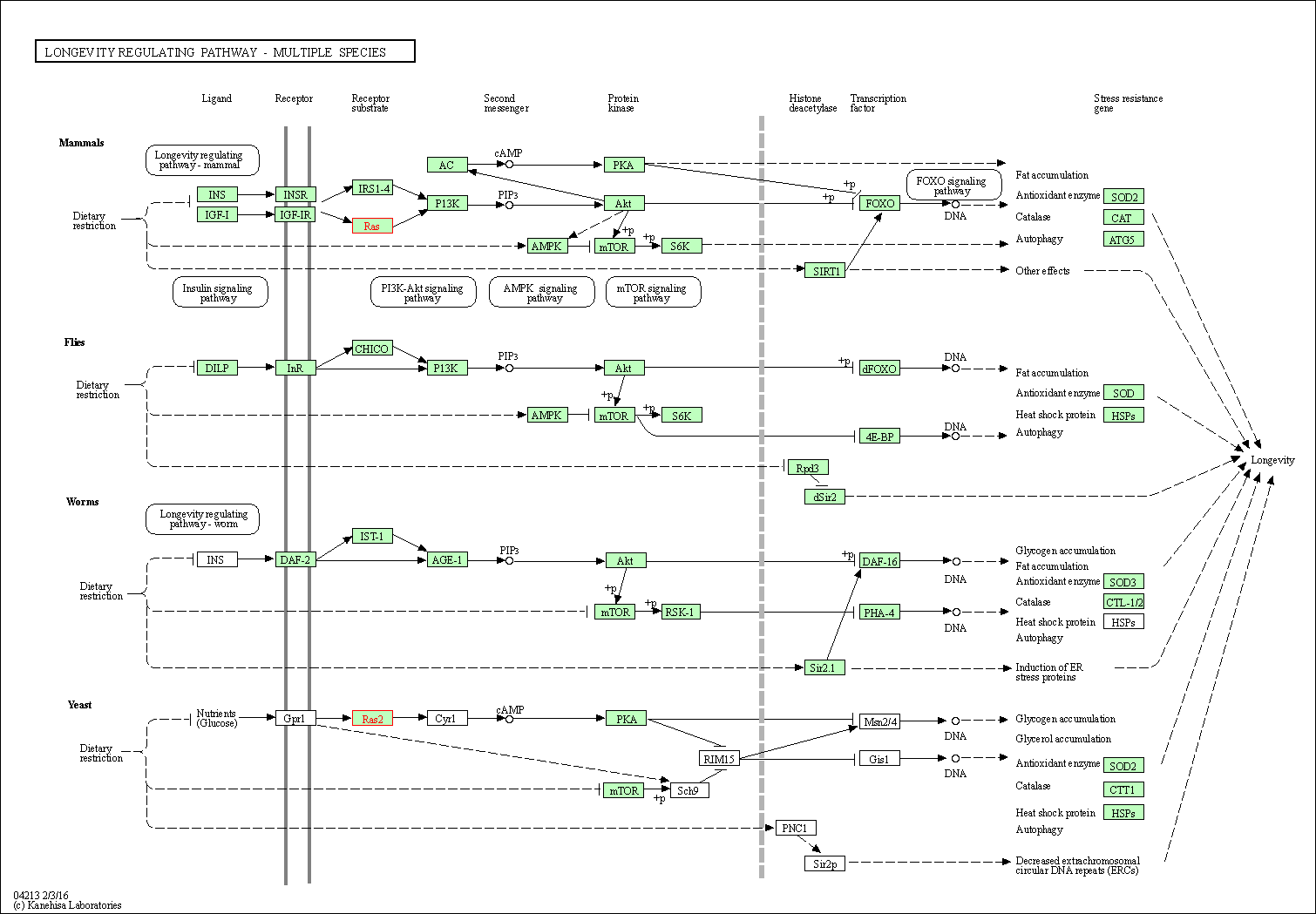
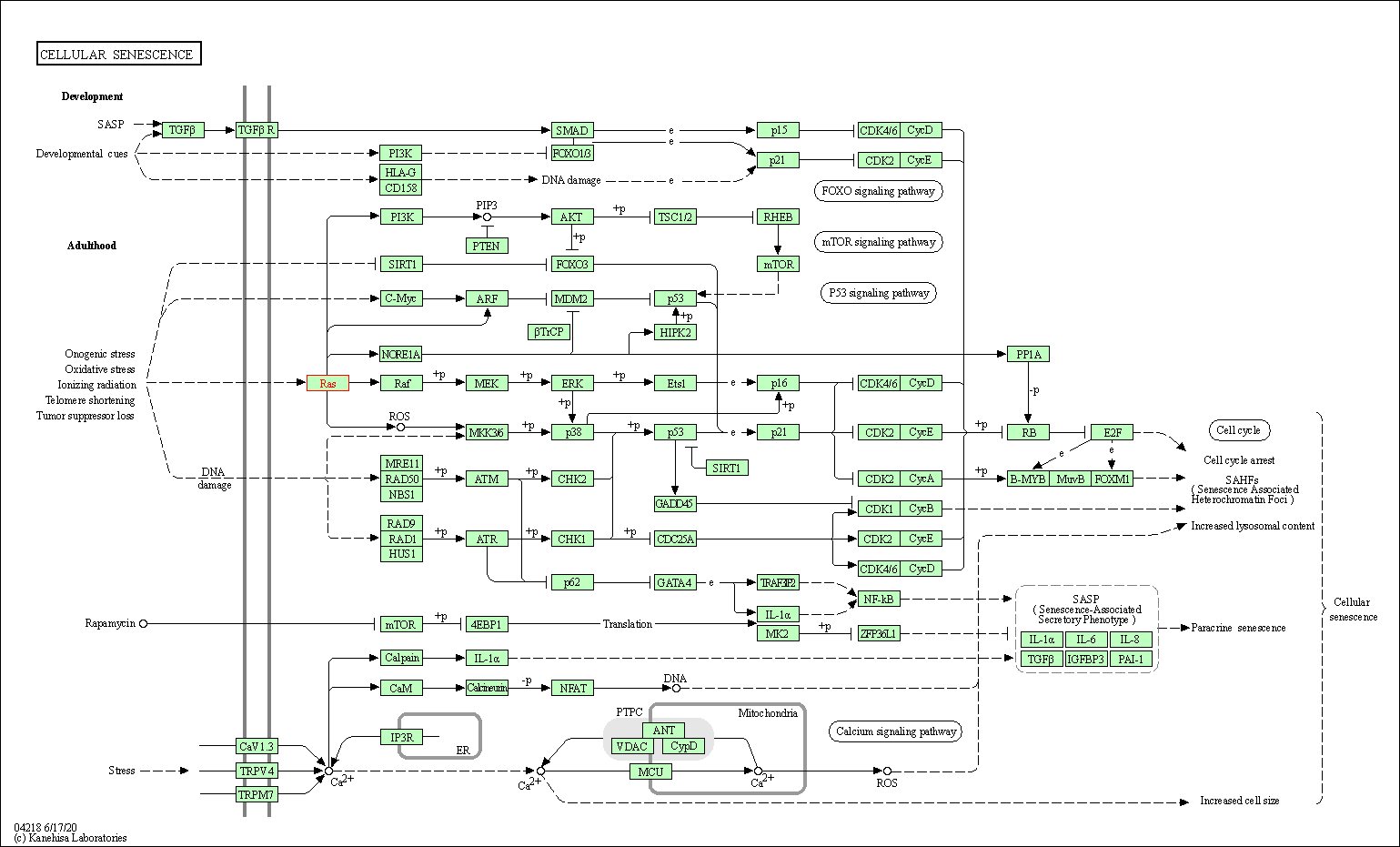
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| MAPK signaling pathway | hsa04010 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| ErbB signaling pathway | hsa04012 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Ras signaling pathway | hsa04014 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Rap1 signaling pathway | hsa04015 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Chemokine signaling pathway | hsa04062 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| FoxO signaling pathway | hsa04068 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Sphingolipid signaling pathway | hsa04071 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Phospholipase D signaling pathway | hsa04072 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
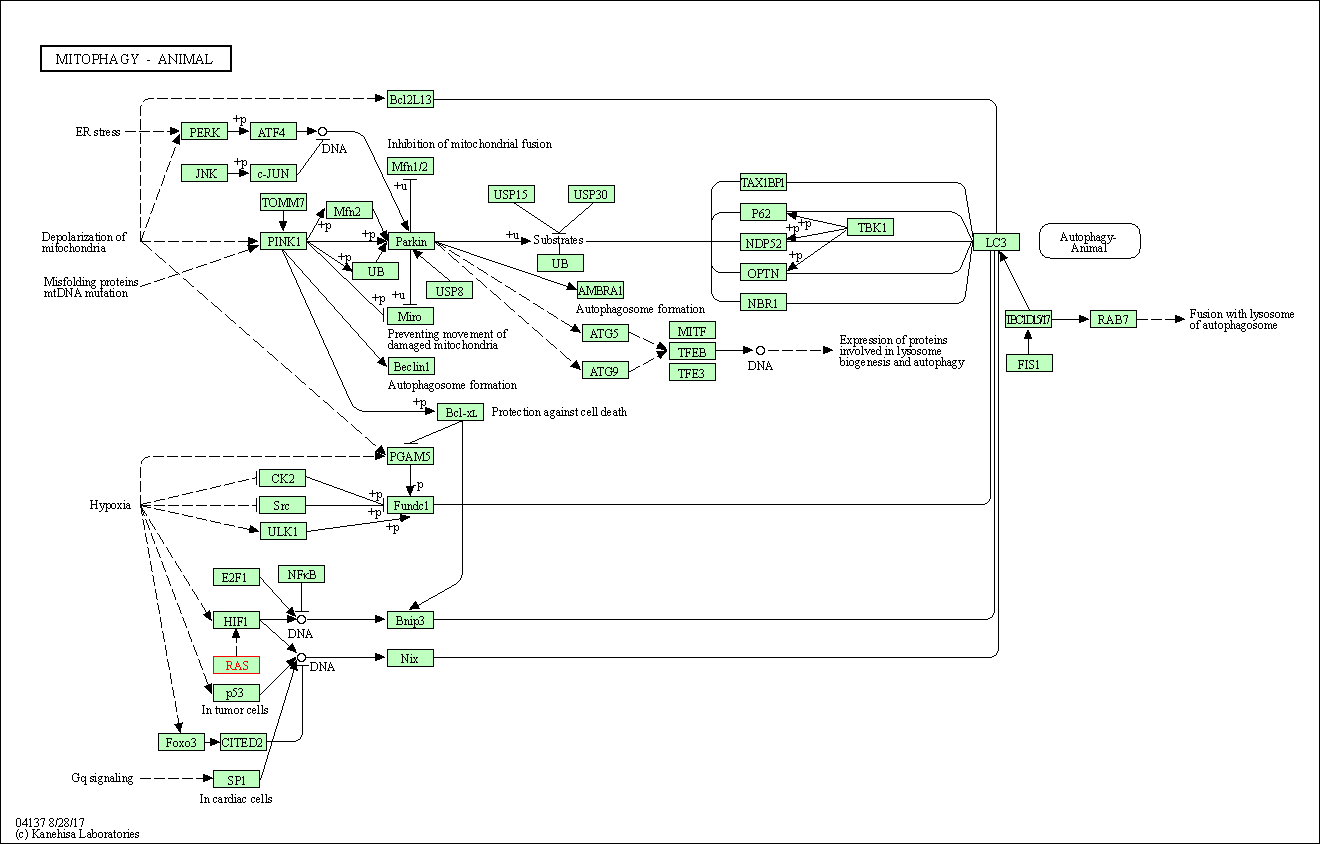
| Mitophagy - animal | hsa04137 | Affiliated Target |

|
| Class: Cellular Processes => Transport and catabolism | Pathway Hierarchy | ||
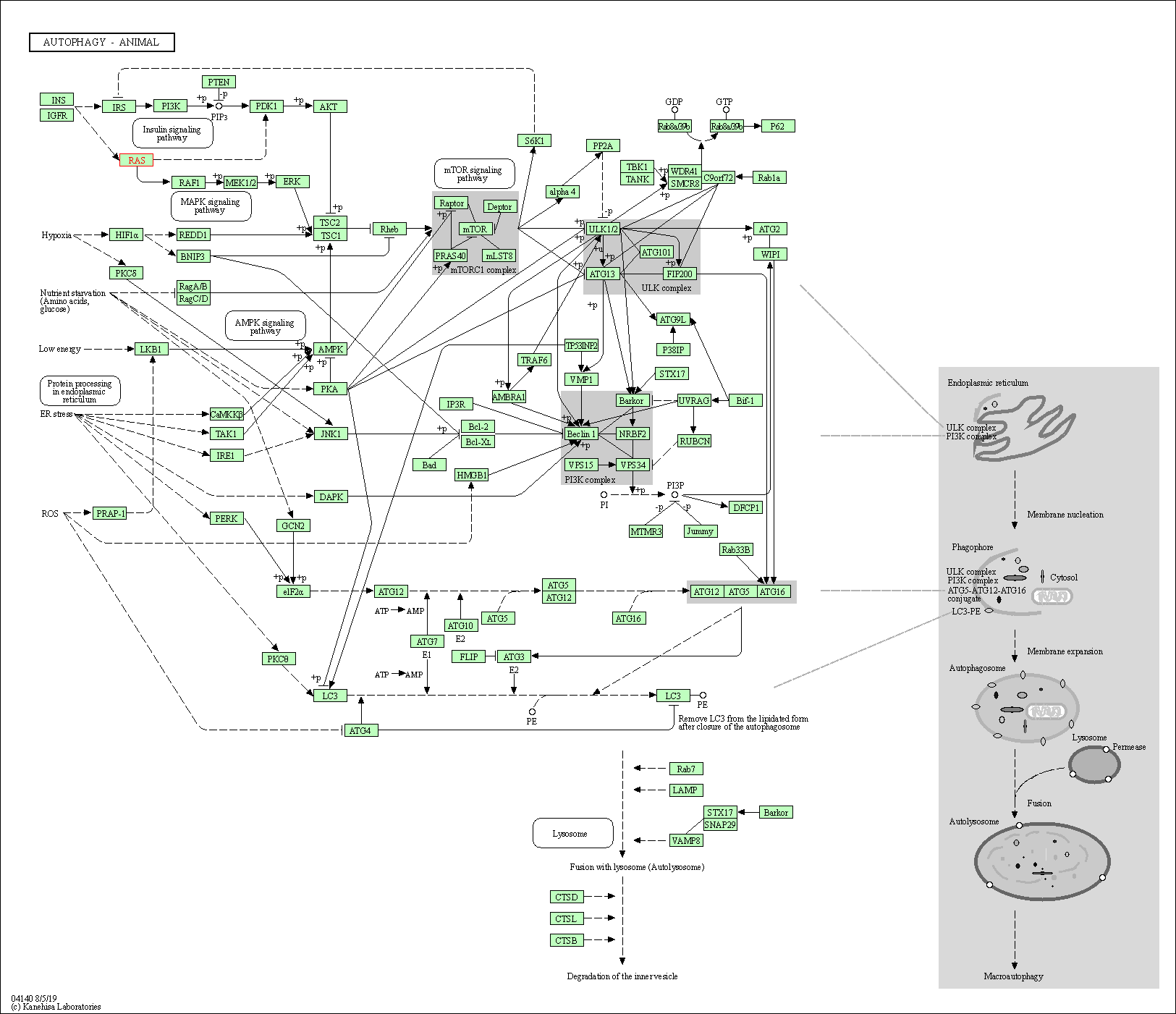
| Autophagy - animal | hsa04140 | Affiliated Target |

|
| Class: Cellular Processes => Transport and catabolism | Pathway Hierarchy | ||
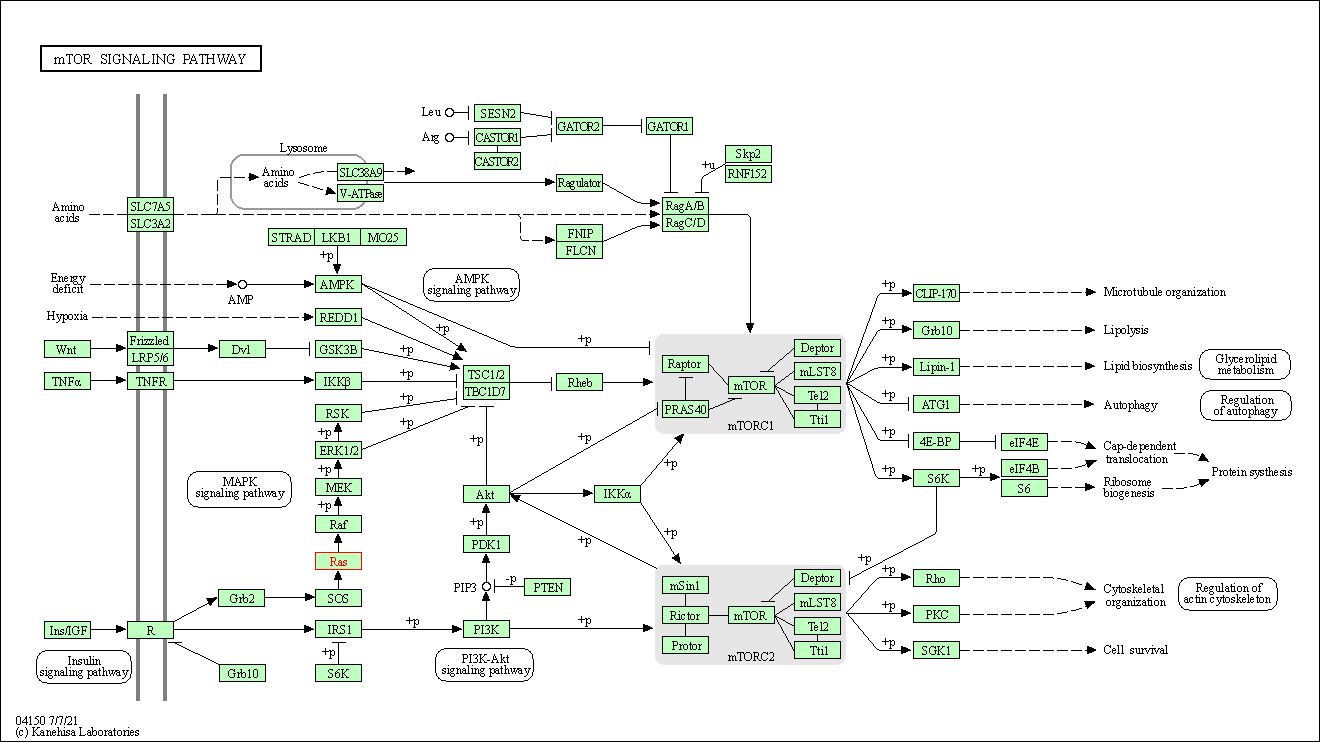
| mTOR signaling pathway | hsa04150 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
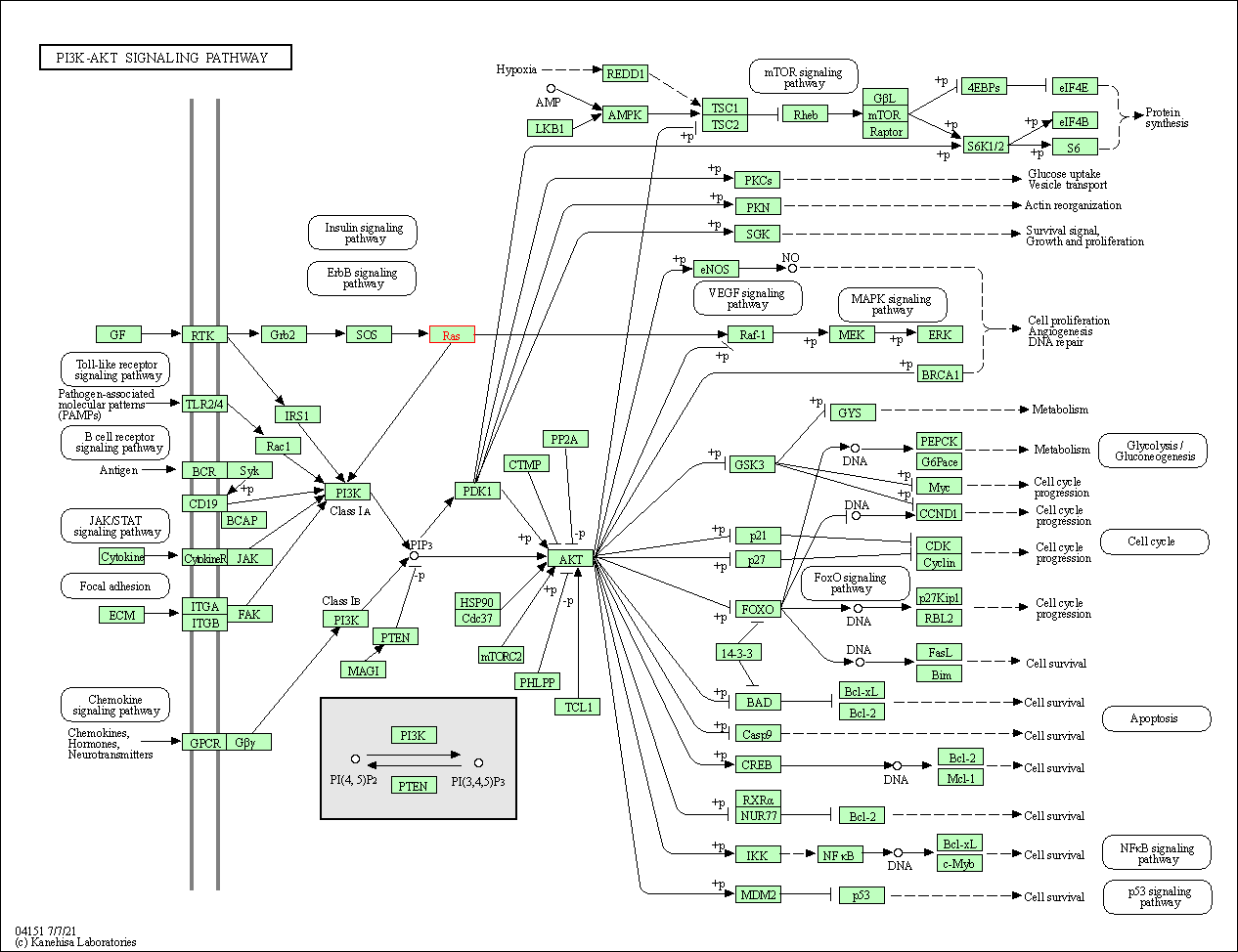
| PI3K-Akt signaling pathway | hsa04151 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Apoptosis | hsa04210 | Affiliated Target |

|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Longevity regulating pathway | hsa04211 | Affiliated Target |

|
| Class: Organismal Systems => Aging | Pathway Hierarchy | ||
| Longevity regulating pathway - multiple species | hsa04213 | Affiliated Target |

|
| Class: Organismal Systems => Aging | Pathway Hierarchy | ||
| Cellular senescence | hsa04218 | Affiliated Target |

|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
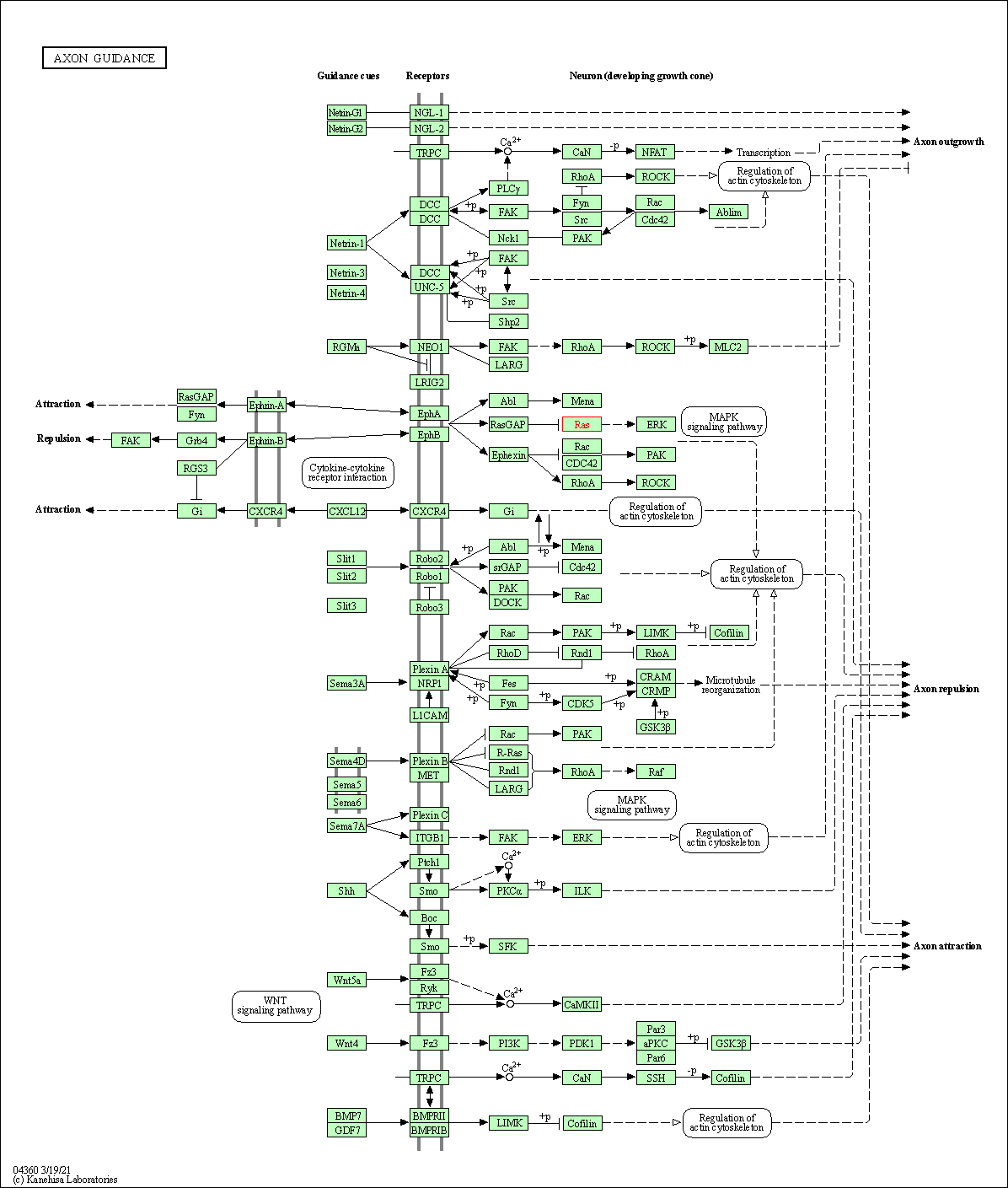
| Axon guidance | hsa04360 | Affiliated Target |

|
| Class: Organismal Systems => Development and regeneration | Pathway Hierarchy | ||
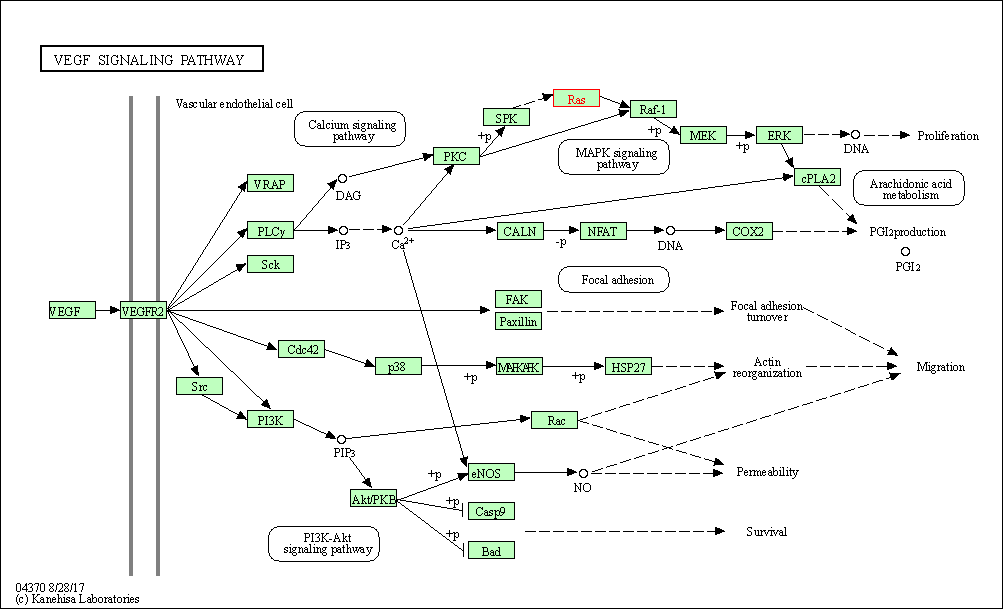
| VEGF signaling pathway | hsa04370 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
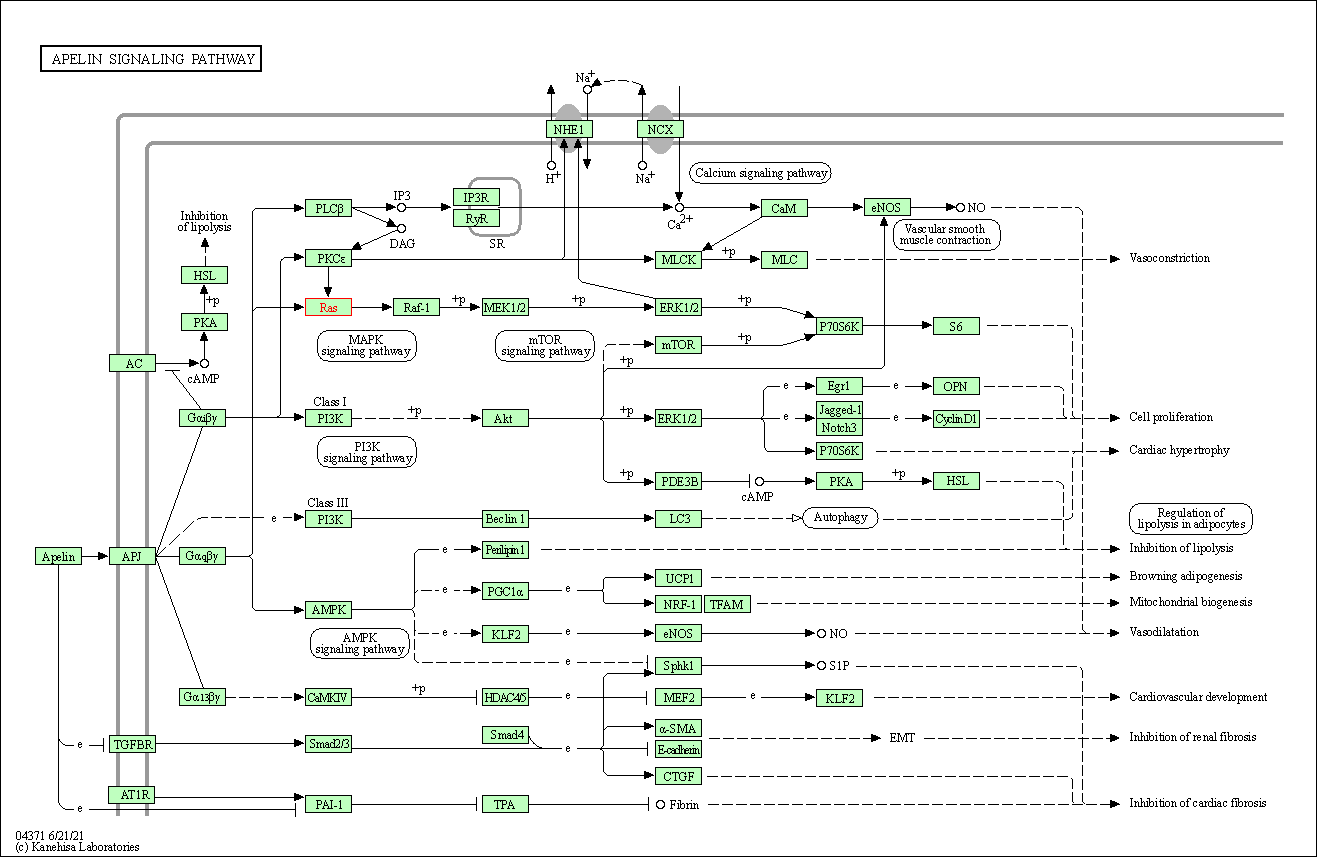
| Apelin signaling pathway | hsa04371 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
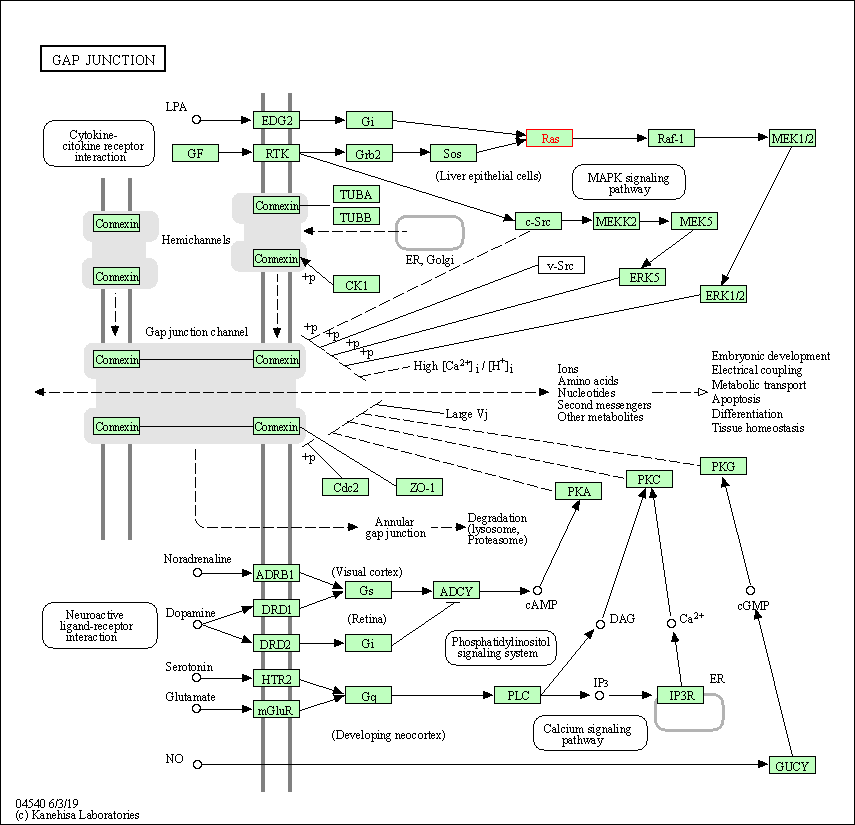
| Gap junction | hsa04540 | Affiliated Target |

|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
| Signaling pathways regulating pluripotency of stem cells | hsa04550 | Affiliated Target |

|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
| C-type lectin receptor signaling pathway | hsa04625 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Natural killer cell mediated cytotoxicity | hsa04650 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| T cell receptor signaling pathway | hsa04660 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
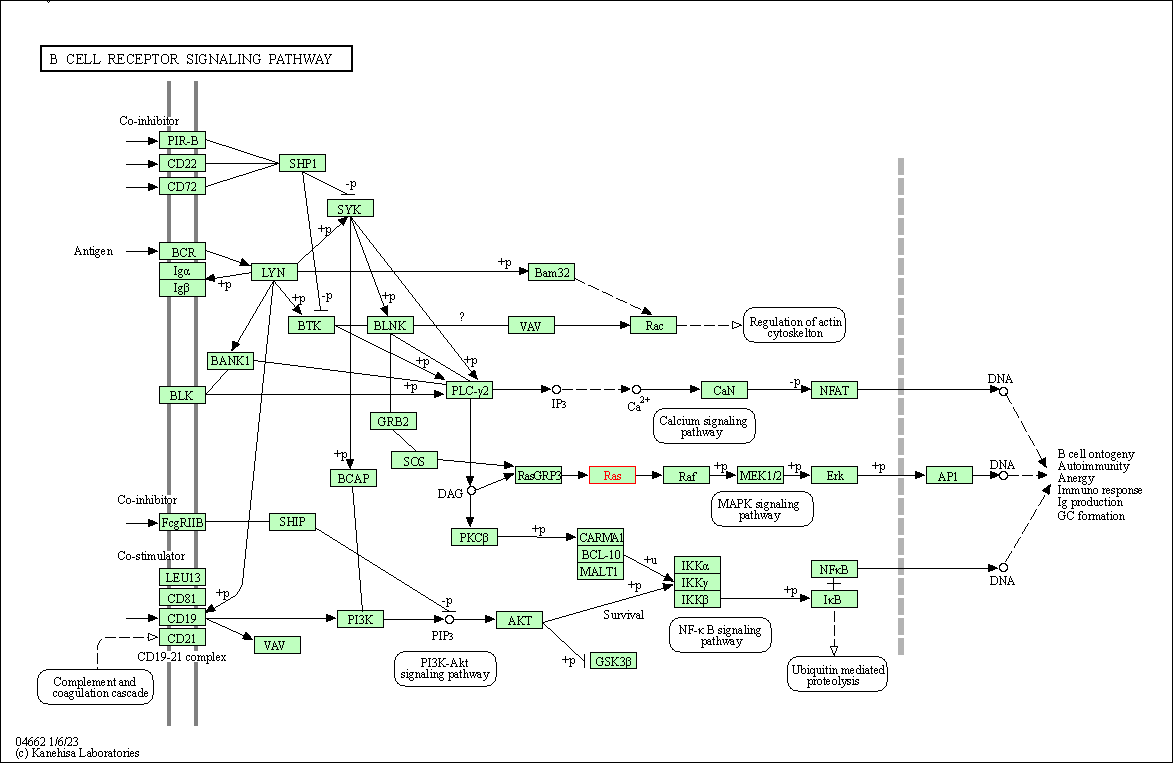
| B cell receptor signaling pathway | hsa04662 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
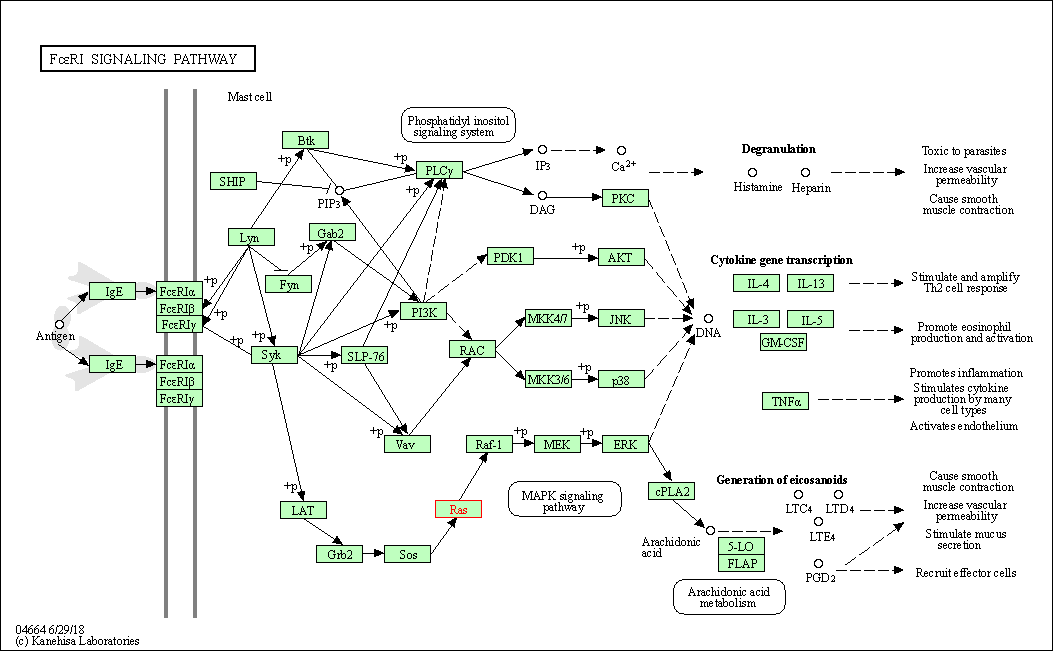
| Fc epsilon RI signaling pathway | hsa04664 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
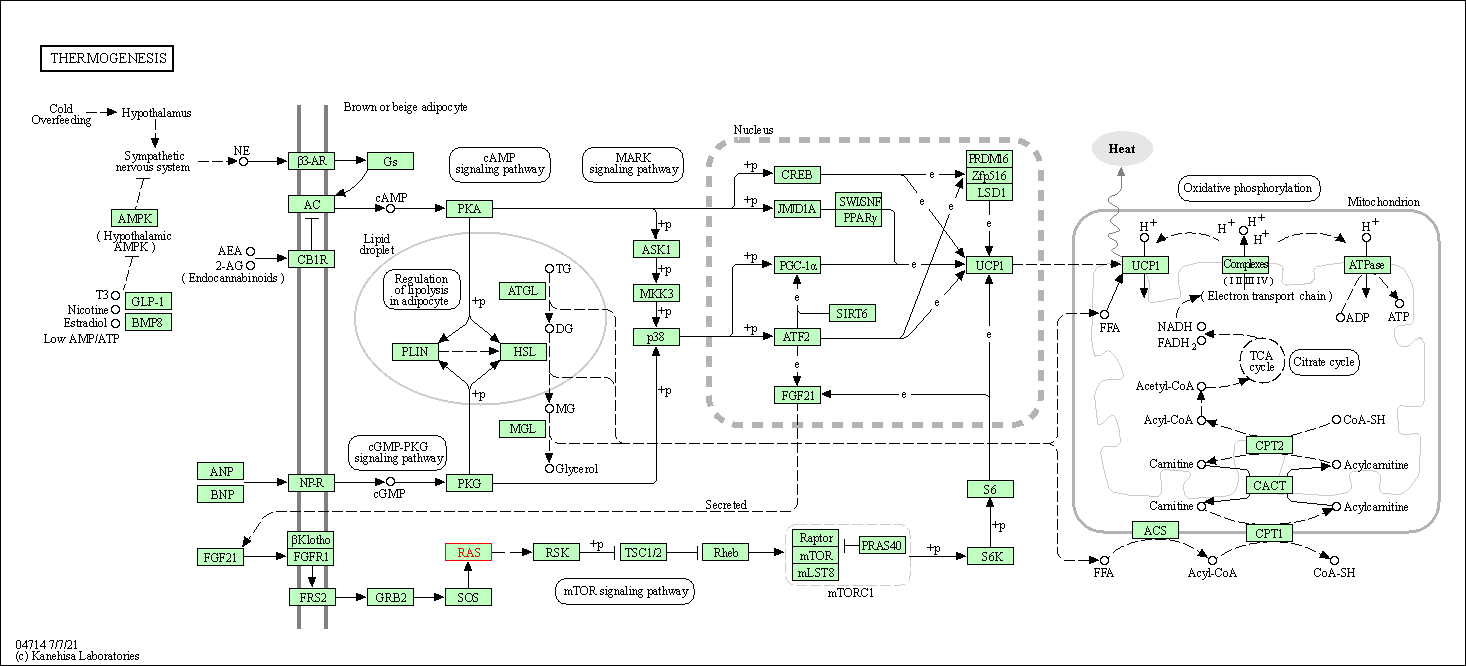
| Thermogenesis | hsa04714 | Affiliated Target |

|
| Class: Organismal Systems => Environmental adaptation | Pathway Hierarchy | ||
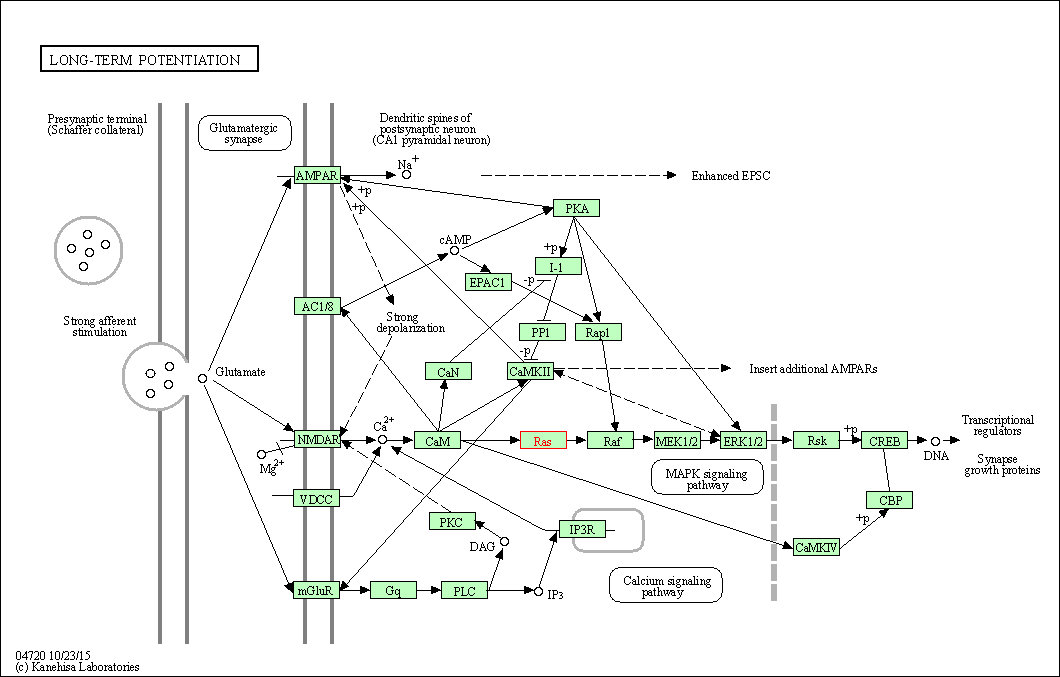
| Long-term potentiation | hsa04720 | Affiliated Target |

|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Neurotrophin signaling pathway | hsa04722 | Affiliated Target |

|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Cholinergic synapse | hsa04725 | Affiliated Target |

|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Serotonergic synapse | hsa04726 | Affiliated Target |

|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Long-term depression | hsa04730 | Affiliated Target |

|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Regulation of actin cytoskeleton | hsa04810 | Affiliated Target |

|
| Class: Cellular Processes => Cell motility | Pathway Hierarchy | ||
| Insulin signaling pathway | hsa04910 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| GnRH signaling pathway | hsa04912 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Progesterone-mediated oocyte maturation | hsa04914 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Estrogen signaling pathway | hsa04915 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Melanogenesis | hsa04916 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Prolactin signaling pathway | hsa04917 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Thyroid hormone signaling pathway | hsa04919 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Oxytocin signaling pathway | hsa04921 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Relaxin signaling pathway | hsa04926 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| GnRH secretion | hsa04929 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Growth hormone synthesis, secretion and action | hsa04935 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Aldosterone-regulated sodium reabsorption | hsa04960 | Affiliated Target |

|
| Class: Organismal Systems => Excretory system | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Clinical pipeline report, company report or official report of Amgen. | |||||
| REF 2 | FDA Approved Drug Products from FDA Official Website. 2022. Application Number: 216340. | |||||
| REF 3 | FDA Approved Drug Products from FDA Official Website. 2023. Application Number: 214665. | |||||
| REF 4 | ClinicalTrials.gov (NCT05132075) A Randomized, Controlled, Open Label, Phase III Study Evaluating the Efficacy and Safety of JDQ443 Versus Docetaxel in Previously Treated Subjects With Locally Advanced or Metastatic KRAS G12C Mutant Non-small Cell Lung Cancer. U.S.National Institutes of Health. | |||||
| REF 5 | ClinicalTrials.gov (NCT04165031) A Study of LY3499446 in Participants With Advanced Solid Tumors With KRAS G12C Mutation. U.S. National Institutes of Health. | |||||
| REF 6 | ClinicalTrials.gov (NCT04006301) First-in-Human Study of JNJ-74699157 in Participants With Tumors Harboring the KRAS G12C Mutation. U.S. National Institutes of Health. | |||||
| REF 7 | Clinical pipeline report, company report or official report of Roche. | |||||
| REF 8 | Clinical pipeline report, company report or official report of Mirati Therapeutics. | |||||
| REF 9 | Discovery, Preclinical Characterization, and Early Clinical Activity of JDQ443, a Structurally Novel, Potent, and Selective Covalent Oral Inhibitor of KRASG12C. Cancer Discov. 2022 Jun 2;12(6):1500-1517. | |||||
| REF 10 | KRAS: From undruggable to a druggable Cancer Target. Cancer Treat Rev. 2020 Sep;89:102070. | |||||
| REF 11 | Fragment Optimization of Reversible Binding to the Switch II Pocket on KRAS Leads to a Potent, In Vivo Active KRAS(G12C) Inhibitor. J Med Chem. 2022 Nov 10;65(21):14614-14629. | |||||
| REF 12 | K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013 Nov 28;503(7477):548-51. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

