Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T33641
(Former ID: TTDI02047)
|
|||||
| Target Name |
Cholesterol 24-hydroxylase (CYP46A1)
|
|||||
| Synonyms |
Cytochrome P450 46A1; CYP46; Cholesterol 24S-hydroxylase; Cholesterol 24-monooxygenase; CH24H
Click to Show/Hide
|
|||||
| Gene Name |
CYP46A1
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 4 Target-related Diseases | + | ||||
| 1 | Epilepsy/seizure [ICD-11: 8A61-8A6Z] | |||||
| 2 | Epileptic encephalopathy [ICD-11: 8A62] | |||||
| 3 | Phakomatoses/hamartoneoplastic syndrome [ICD-11: LD2D] | |||||
| 4 | Acidosis [ICD-11: 5C73] | |||||
| Function |
Primarily catalyzes the hydroxylation (with S stereochemistry) at C-24 of cholesterol side chain, triggering cholesterol diffusion out of neurons and its further degradation. By promoting constant cholesterol elimination in neurons, may activate the mevalonate pathway and coordinate the synthesis of new cholesterol and nonsterol isoprenoids involved in synaptic activity and learning. Further hydroxylates cholesterol derivatives and hormone steroids on both the ring and side chain of these molecules, converting them into active oxysterols involved in lipid signaling and biosynthesis. Acts as an epoxidase converting cholesta-5,24-dien-3beta-ol/desmosterol into (24S),25-epoxycholesterol, an abundant lipid ligand of nuclear NR1H2 and NR1H3 receptors shown to promote neurogenesis in developing brain. May also catalyze the oxidative metabolism of xenobiotics, such as clotrimazole. P450 monooxygenase that plays a major role in cholesterol homeostasis in the brain.
Click to Show/Hide
|
|||||
| BioChemical Class |
Paired donor oxygen oxidoreductase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 1.14.14.25
|
|||||
| Sequence |
MSPGLLLLGSAVLLAFGLCCTFVHRARSRYEHIPGPPRPSFLLGHLPCFWKKDEVGGRVL
QDVFLDWAKKYGPVVRVNVFHKTSVIVTSPESVKKFLMSTKYNKDSKMYRALQTVFGERL FGQGLVSECNYERWHKQRRVIDLAFSRSSLVSLMETFNEKAEQLVEILEAKADGQTPVSM QDMLTYTAMDILAKAAFGMETSMLLGAQKPLSQAVKLMLEGITASRNTLAKFLPGKRKQL REVRESIRFLRQVGRDWVQRRREALKRGEEVPADILTQILKAEEGAQDDEGLLDNFVTFF IAGHETSANHLAFTVMELSRQPEIVARLQAEVDEVIGSKRYLDFEDLGRLQYLSQVLKES LRLYPPAWGTFRLLEEETLIDGVRVPGNTPLLFSTYVMGRMDTYFEDPLTFNPDRFGPGA PKPRFTYFPFSLGHRSCIGQQFAQMEVKVVMAKLLQRLEFRLVPGQRFGLQEQATLKPLD PVLCTLRPRGWQPAPPPPPC Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| HIT2.0 ID | T44X5B | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 1 Clinical Trial Drugs | + | ||||
| 1 | TAK-935 | Drug Info | Phase 2 | Dravet syndrome | [2] | |
| Mode of Action | [+] 1 Modes of Action | + | ||||
| Inhibitor | [+] 11 Inhibitor drugs | + | ||||
| 1 | TAK-935 | Drug Info | [1] | |||
| 2 | PMID25514969-Compound-10 | Drug Info | [3] | |||
| 3 | PMID25514969-Compound-11 | Drug Info | [3] | |||
| 4 | PMID25514969-Compound-12 | Drug Info | [3] | |||
| 5 | PMID25514969-Compound-13 | Drug Info | [3] | |||
| 6 | PMID25514969-Compound-8 | Drug Info | [3] | |||
| 7 | PMID25514969-Compound-9 | Drug Info | [3] | |||
| 8 | PMID25514969-Compound-Figure2-2 | Drug Info | [3] | |||
| 9 | PMID25514969-Compound-Figure2-3 | Drug Info | [3] | |||
| 10 | PMID25514969-Compound-Figure2-4 | Drug Info | [3] | |||
| 11 | PMID25514969-Compound-Figure3 | Drug Info | [3] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Tranylcypromine | Ligand Info | |||||
| Structure Description | Tranylcypromine complex of Cytochrome P450 46A1 | PDB:3MDR | ||||
| Method | X-ray diffraction | Resolution | 2.00 Å | Mutation | No | [4] |
| PDB Sequence |
RVLQDVFLDW
67 AKKYGPVVRV77 NVFHKTSVIV87 TSPESVKKFL97 MSTKYNKDSK107 MYRALQTVFG 117 ERLFGQGLVS127 ECNYERWHKQ137 RRVIDLAFSR147 SSLVSLMETF157 NEKAEQLVEI 167 LEAKADGQTP177 VSMQDMLTYT187 AMDILAKAAF197 GMETSMLLGA207 QKPLSQAVKL 217 MLEGITASRN227 TKRKQLREVR244 ESIRFLRQVG254 RDWVQRRREA264 LKRGEEVPAD 274 ILTQILKAEE284 GAQDDEGLLD294 NFVTFFIAGH304 ETSANHLAFT314 VMELSRQPEI 324 VARLQAEVDE334 VIGSKRYLDF344 EDLGRLQYLS354 QVLKESLRLY364 PPAWGTFRLL 374 EEETLIDGVR384 VPGNTPLLFS394 TYVMGRMDTY404 FEDPLTFNPD414 RFGPGAPKPR 424 FTYFPFSLGH434 RSCIGQQFAQ444 MEVKVVMAKL454 LQRLEFRLVP464 GQRFGLQEQA 474 TLKPLDPVLC484 TLRPRGW
|
|||||
|
|
||||||
| Ligand Name: Voriconazole | Ligand Info | |||||
| Structure Description | Voriconazole complex of Cytochrome P450 46A1 | PDB:3MDT | ||||
| Method | X-ray diffraction | Resolution | 2.30 Å | Mutation | No | [4] |
| PDB Sequence |
VLQDVFLDWA
68 KKYGPVVRVN78 VFHKTSVIVT88 SPESVKKFLM98 STKYNKDSKM108 YRALQTVFGE 118 RLFGQGLVSE128 CNYERWHKQR138 RVIDLAFSRS148 SLVSLMETFN158 EKAEQLVEIL 168 EAKADGQTPV178 SMQDMLTYTA188 MDILAKAAFG198 METSMLLGAQ208 KPLSQAVKLM 218 LEGITASRNT228 KRKQLREVRE245 SIRFLRQVGR255 DWVQRRREAL265 KRGEEVPADI 275 LTQILKAEEG285 AQDDEGLLDN295 FVTFFIAGHE305 TSANHLAFTV315 MELSRQPEIV 325 ARLQAEVDEV335 IGSKRYLDFE345 DLGRLQYLSQ355 VLKESLRLYP365 PAWGTFRLLE 375 EETLIDGVRV385 PGNTPLLFST395 YVMGRMDTYF405 EDPLTFNPDR415 FGPGAPKPRF 425 TYFPFSLGHR435 SCIGQQFAQM445 EVKVVMAKLL455 QRLEFRLVPG465 QRFGLQEQAT 475 LKPLDPVLCT485 LRPR
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
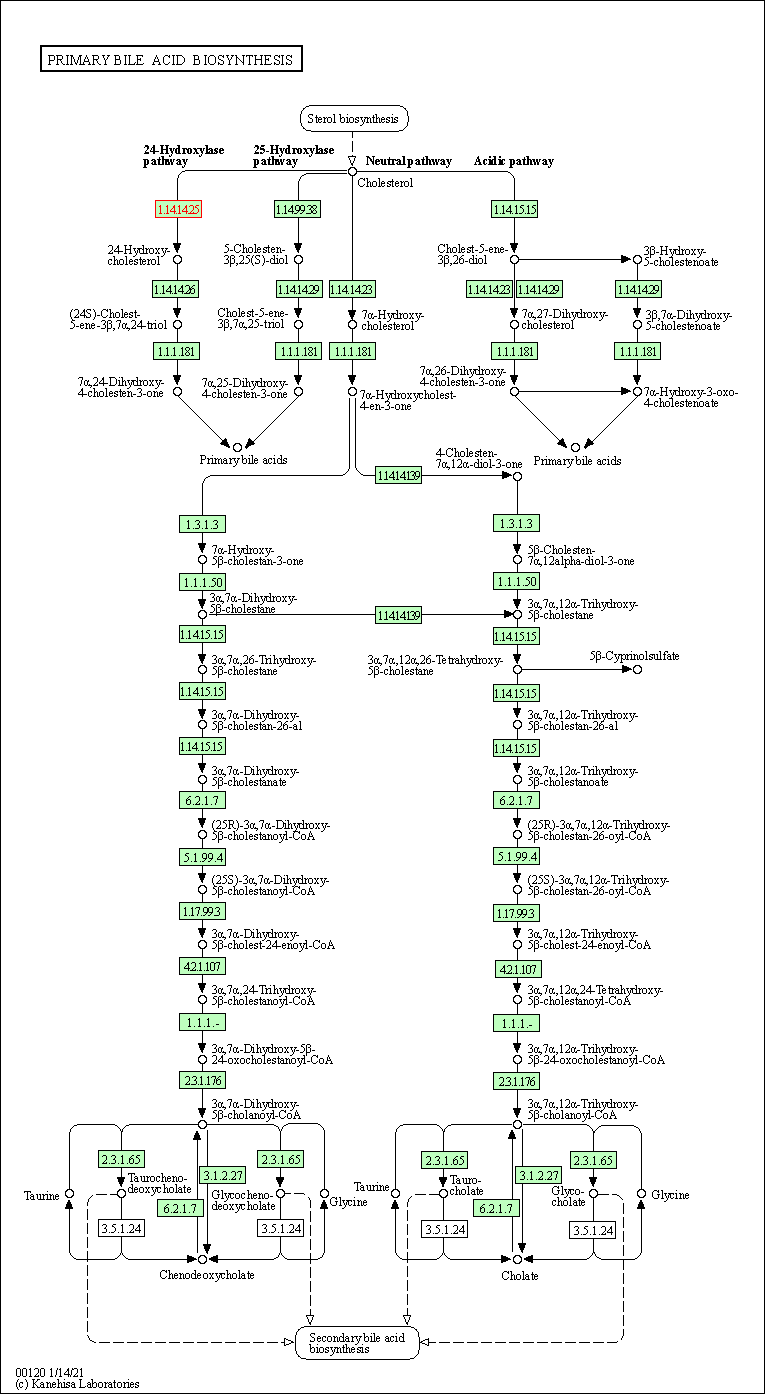
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Primary bile acid biosynthesis | hsa00120 | Affiliated Target |

|
| Class: Metabolism => Lipid metabolism | Pathway Hierarchy | ||
| Degree | 3 | Degree centrality | 3.22E-04 | Betweenness centrality | 9.97E-05 |
|---|---|---|---|---|---|
| Closeness centrality | 1.67E-01 | Radiality | 1.26E+01 | Clustering coefficient | 3.33E-01 |
| Neighborhood connectivity | 5.00E+00 | Topological coefficient | 4.67E-01 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 1 KEGG Pathways | + | ||||
| 1 | Primary bile acid biosynthesis | |||||
| Pathwhiz Pathway | [+] 1 Pathwhiz Pathways | + | ||||
| 1 | Bile Acid Biosynthesis | |||||
| Reactome | [+] 1 Reactome Pathways | + | ||||
| 1 | Endogenous sterols | |||||
| WikiPathways | [+] 4 WikiPathways | + | ||||
| 1 | Metapathway biotransformation | |||||
| 2 | Oxidation by Cytochrome P450 | |||||
| 3 | Phase 1 - Functionalization of compounds | |||||
| 4 | Bile acid and bile salt metabolism | |||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800040983) | |||||
| REF 2 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 3 | Imidazo[1,2-a]pyridines as cholesterol 24-hydroxylase (CYP46A1) inhibitors: a patent evaluation (WO2014061676).Expert Opin Ther Pat. 2015 Mar;25(3):373-7. | |||||
| REF 4 | Structural basis of drug binding to CYP46A1, an enzyme that controls cholesterol turnover in the brain. J Biol Chem. 2010 Oct 8;285(41):31783-95. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

