Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T18664
(Former ID: TTDI02646)
|
|||||
| Target Name |
Monoglyceride lipase (MAGL)
|
|||||
| Synonyms |
Monoacylglycerol lipase; MGL; Lysophospholipaselike; Lysophospholipase-like; Lysophospholipase homolog; HUK5; HU-K5
Click to Show/Hide
|
|||||
| Gene Name |
MGLL
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 2 Target-related Diseases | + | ||||
| 1 | General pain disorder [ICD-11: 8E43] | |||||
| 2 | Tic disorder [ICD-11: 8A05] | |||||
| Function |
Hydrolyzes the endocannabinoid 2-arachidonoylglycerol, and thereby contributes to the regulation of endocannabinoid signaling, nociperception and perception of pain. Regulates the levels of fatty acids that serve as signaling molecules and promote cancer cell migration, invasion and tumor growth. Converts monoacylglycerides to free fatty acids and glycerol.
Click to Show/Hide
|
|||||
| BioChemical Class |
Carboxylic ester hydrolase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 3.1.1.23
|
|||||
| Sequence |
MPEESSPRRTPQSIPYQDLPHLVNADGQYLFCRYWKPTGTPKALIFVSHGAGEHSGRYEE
LARMLMGLDLLVFAHDHVGHGQSEGERMVVSDFHVFVRDVLQHVDSMQKDYPGLPVFLLG HSMGGAIAILTAAERPGHFAGMVLISPLVLANPESATTFKVLAAKVLNLVLPNLSLGPID SSVLSRNKTEVDIYNSDPLICRAGLKVCFGIQLLNAVSRVERALPKLTVPFLLLQGSADR LCDSKGAYLLMELAKSQDKTLKIYEGAYHVLHKELPEVTNSVFHEINMWVSQRTATAGTA SPP Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| HIT2.0 ID | T95CAO | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 1 Clinical Trial Drugs | + | ||||
| 1 | ABX-1431 | Drug Info | Phase 1 | Neuropathic pain | [1] | |
| Mode of Action | [+] 1 Modes of Action | + | ||||
| Inhibitor | [+] 47 Inhibitor drugs | + | ||||
| 1 | ABX-1431 | Drug Info | [1] | |||
| 2 | Azetidine-benzoxazin-3(4H)-one derivative 1 | Drug Info | [2] | |||
| 3 | Azetidine-benzoxazin-3(4H)-one derivative 2 | Drug Info | [2] | |||
| 4 | Azetidine-benzoxazin-3(4H)-one derivative 3 | Drug Info | [2] | |||
| 5 | Azetidinyl-piperazine derivative 1 | Drug Info | [2] | |||
| 6 | Azetidinyl-piperazine derivative 2 | Drug Info | [2] | |||
| 7 | Azetidinyl-piperazine derivative 3 | Drug Info | [2] | |||
| 8 | Azetidinyl-piperidine derivative 1 | Drug Info | [2] | |||
| 9 | Azetidinyl-piperidine derivative 2 | Drug Info | [2] | |||
| 10 | Azetidinyl-piperidine derivative 3 | Drug Info | [2] | |||
| 11 | Carbamate derivative 10 | Drug Info | [2] | |||
| 12 | Carbamate derivative 11 | Drug Info | [2] | |||
| 13 | Carbamate derivative 12 | Drug Info | [2] | |||
| 14 | Carbamate derivative 13 | Drug Info | [2] | |||
| 15 | Carbamate derivative 14 | Drug Info | [2] | |||
| 16 | Carbamate derivative 15 | Drug Info | [2] | |||
| 17 | Carbamate derivative 16 | Drug Info | [2] | |||
| 18 | Carbamate derivative 17 | Drug Info | [2] | |||
| 19 | Carbamate derivative 9 | Drug Info | [2] | |||
| 20 | Carbamide derivative 24 | Drug Info | [2] | |||
| 21 | Carbamide derivative 25 | Drug Info | [2] | |||
| 22 | Carbamide derivative 26 | Drug Info | [2] | |||
| 23 | Piperazine carbamic compound 1 | Drug Info | [2] | |||
| 24 | Piperazine carbamic compound 2 | Drug Info | [2] | |||
| 25 | Piperazine carbamic compound 3 | Drug Info | [2] | |||
| 26 | Piperazine carbamic compound 4 | Drug Info | [2] | |||
| 27 | Piperazine carbamic compound 5 | Drug Info | [2] | |||
| 28 | PMID29053063-Compound-11c | Drug Info | [2] | |||
| 29 | PMID29053063-Compound-11d | Drug Info | [2] | |||
| 30 | PMID29053063-Compound-14 | Drug Info | [2] | |||
| 31 | PMID29053063-Compound-15 | Drug Info | [2] | |||
| 32 | PMID29053063-Compound-17 | Drug Info | [2] | |||
| 33 | PMID29053063-Compound-4 | Drug Info | [2] | |||
| 34 | PMID29053063-Compound-5 | Drug Info | [2] | |||
| 35 | PMID29053063-Compound-7a | Drug Info | [2] | |||
| 36 | PMID29053063-Compound-7b | Drug Info | [2] | |||
| 37 | PMID29053063-Compound-7c | Drug Info | [2] | |||
| 38 | PMID29053063-Compound-7d | Drug Info | [2] | |||
| 39 | PMID29053063-Compound-7e | Drug Info | [2] | |||
| 40 | PMID29053063-Compound-7f | Drug Info | [2] | |||
| 41 | Pyrazole derivative 80 | Drug Info | [2] | |||
| 42 | Pyrazole derivative 81 | Drug Info | [2] | |||
| 43 | Pyrazole derivative 82 | Drug Info | [2] | |||
| 44 | Pyrazole derivative 83 | Drug Info | [2] | |||
| 45 | Pyrazole derivative 84 | Drug Info | [2] | |||
| 46 | Pyrazole derivative 85 | Drug Info | [2] | |||
| 47 | JZL184 | Drug Info | [3] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: (2-Cyclohexyl-1,3-Benzoxazol-6-Yl){3-[4-(Pyrimidin-2-Yl)piperazin-1-Yl]azetidin-1-Yl}methanone | Ligand Info | |||||
| Structure Description | Crystal Structure of a soluble form of human MGLL in complex with an inhibitor | PDB:3PE6 | ||||
| Method | X-ray diffraction | Resolution | 1.35 Å | Mutation | Yes | [4] |
| PDB Sequence |
PRRTPQSIPY
16 QDLPHLVNAD26 GQYLFCRYWA36 PTGTPKALIF46 VSHGAGEHSG56 RYEELARMLM 66 GLDLLVFAHD76 HVGHGQSEGE86 RMVVSDFHVF96 VRDVLQHVDS106 MQKDYPGLPV 116 FLLGHSMGGA126 IAILTAAERP136 GHFAGMVLIS146 PLVLANPESA156 TTFKVLAAKV 166 LNSVLPNLSS176 GPIDSSVLSR186 NKTEVDIYNS196 DPLICRAGLK206 VCFGIQLLNA 216 VSRVERALPK226 LTVPFLLLQG236 SADRLCDSKG246 AYLLMELAKS256 QDKTLKIYEG 266 AYHVLHKELP276 EVTNSVFHEI286 NMWVSQRTA
|
|||||
|
|
GLY50
2.463
ALA51
1.986
GLY52
4.848
GLU53
2.595
ARG57
3.782
TYR58
4.411
HIS121
2.743
SER122
2.178
MET123
2.072
GLY124
4.147
LEU148
2.856
ALA151
2.474
ASN152
4.302
SER155
3.002
ALA156
2.467
PHE159
2.997
LYS160
4.667
ILE179
2.588
ASP180
4.985
|
|||||
| Ligand Name: (4R)-1-(2'-chloro[1,1'-biphenyl]-3-yl)-4-[4-(1,3-thiazole-2-carbonyl)piperazin-1-yl]pyrrolidin-2-one | Ligand Info | |||||
| Structure Description | Crystal structure of human monoacylglycerol lipase in complex with compound 3l | PDB:5ZUN | ||||
| Method | X-ray diffraction | Resolution | 1.35 Å | Mutation | Yes | [5] |
| PDB Sequence |
PRRTPQSIPY
16 QDLPHLVNAD26 GQYLFCRYWA36 PTGTPKALIF46 VSHGAGEHSG56 RYEELARMLM 66 GLDLLVFAHD76 HVGHGQSEGE86 RMVVSDFHVF96 VRDVLQHVDS106 MQKDYPGLPV 116 FLLGHSMGGA126 IAILTAAERP136 GHFAGMVLIS146 PLVLANPESA156 TTFKVLAAKV 166 LNSVLPNLSS176 GPIDSSVLSR186 NKTEVDIYNS196 DPLICRAGLK206 VCFGIQLLNA 216 VSRVERALPK226 LTVPFLLLQG236 SADRLCDSKG246 AYLLMELAKS256 QDKTLKIYEG 266 AYHVLHKELP276 EVTNSVFHEI286 NMWVSQRTA
|
|||||
|
|
GLY50
3.362
ALA51
2.795
GLU53
3.217
ARG57
2.823
MET88
3.603
HIS121
3.459
SER122
2.879
MET123
2.887
GLY124
4.602
LEU148
4.498
GLY177
3.432
PRO178
3.666
ILE179
3.609
|
|||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
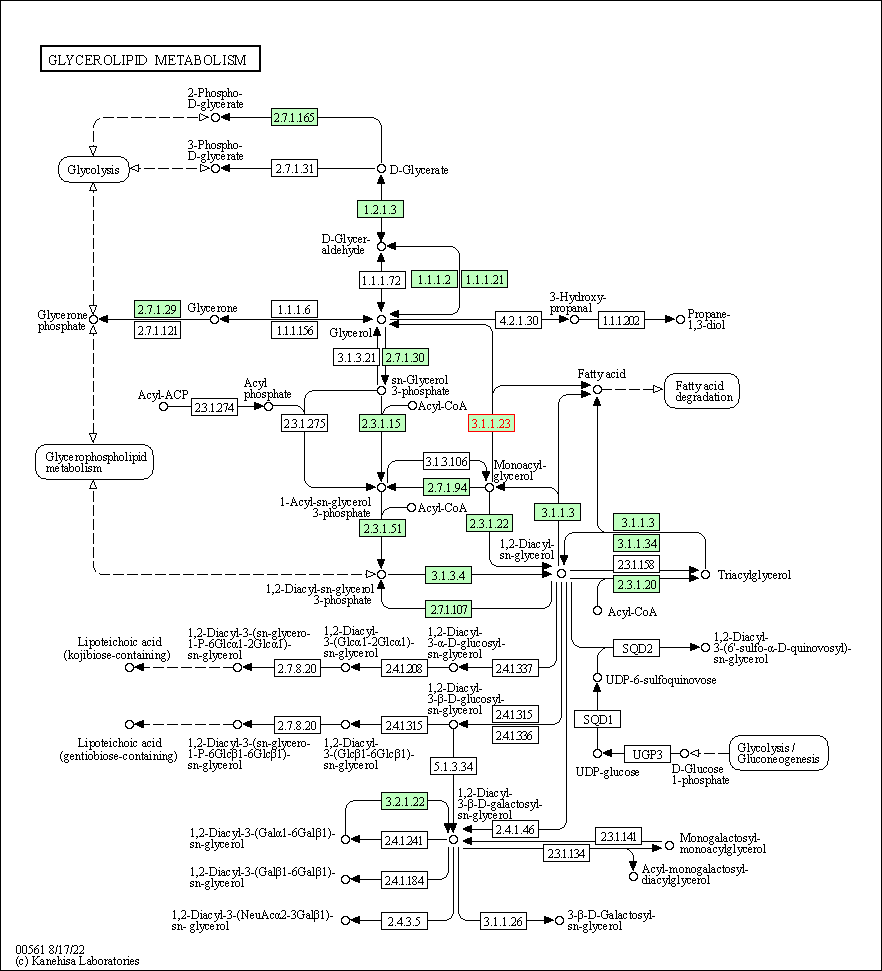
| Glycerolipid metabolism | hsa00561 | Affiliated Target |

|
| Class: Metabolism => Lipid metabolism | Pathway Hierarchy | ||
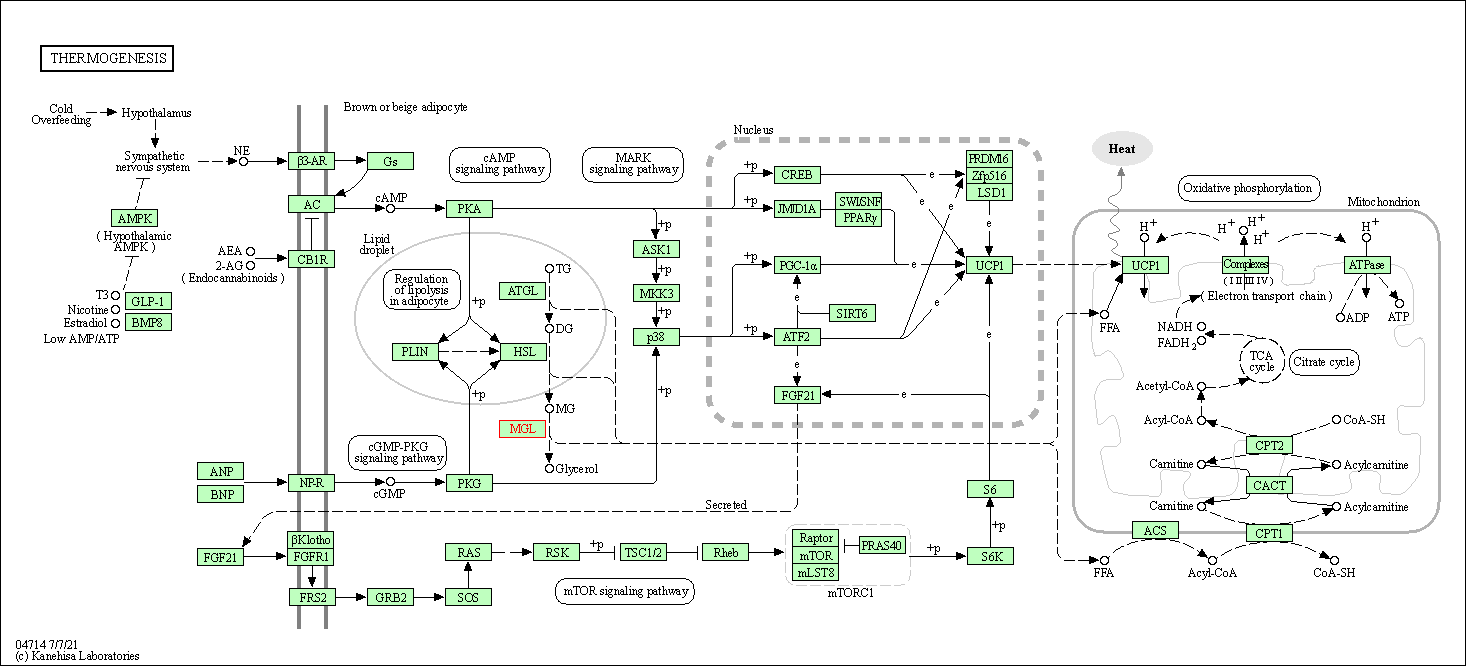
| Thermogenesis | hsa04714 | Affiliated Target |

|
| Class: Organismal Systems => Environmental adaptation | Pathway Hierarchy | ||
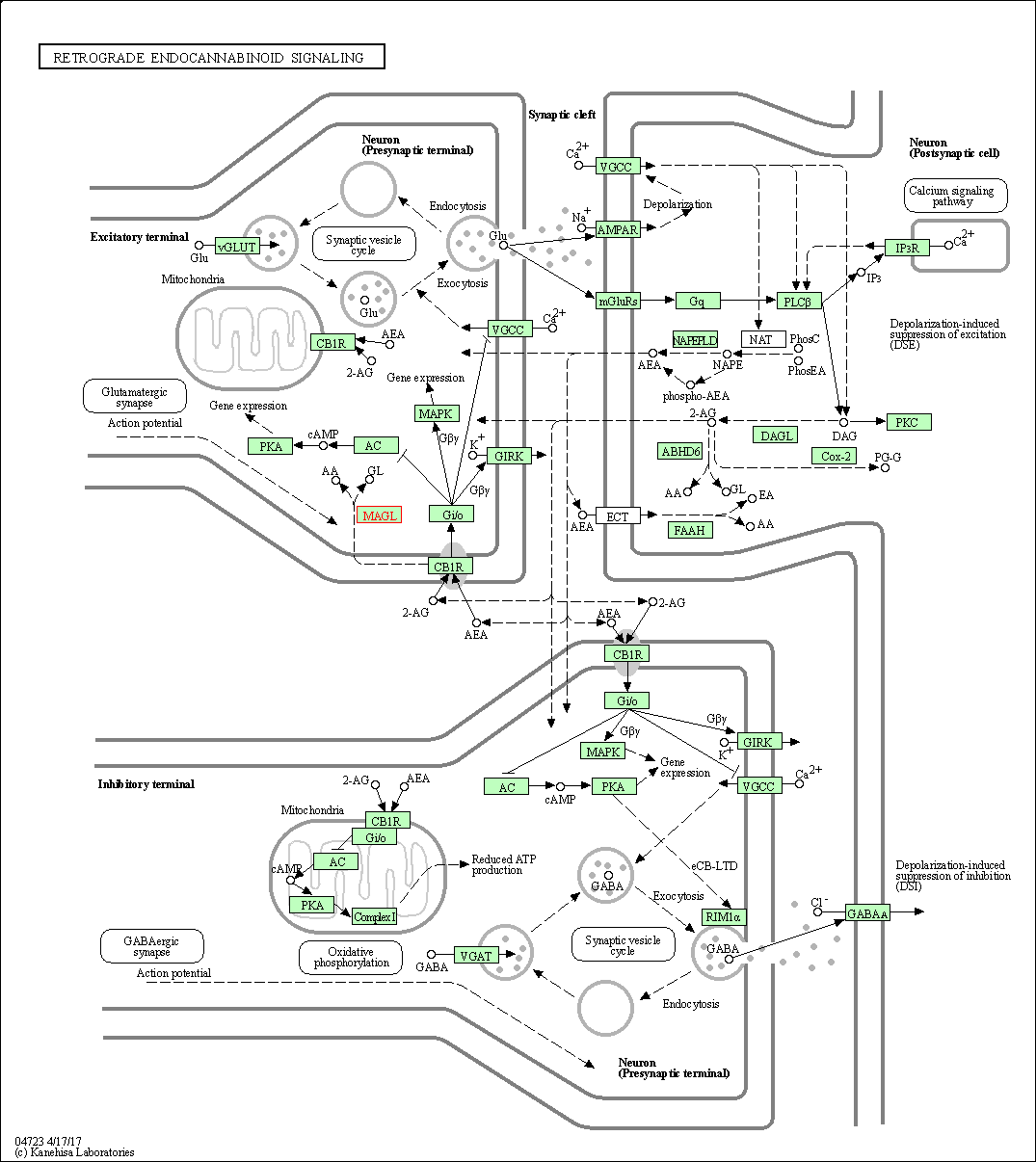
| Retrograde endocannabinoid signaling | hsa04723 | Affiliated Target |

|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
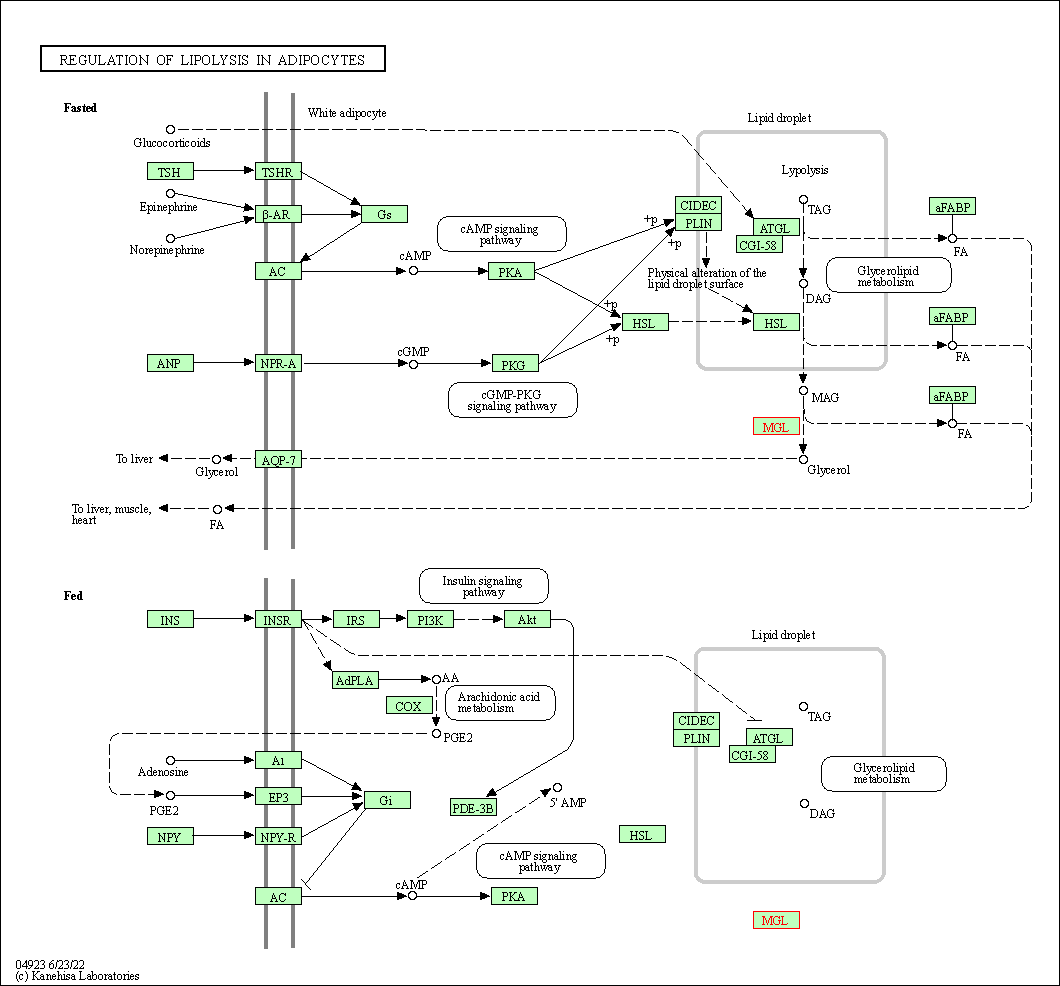
| Regulation of lipolysis in adipocytes | hsa04923 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Degree | 1 | Degree centrality | 1.07E-04 | Betweenness centrality | 0.00E+00 |
|---|---|---|---|---|---|
| Closeness centrality | 1.58E-01 | Radiality | 1.23E+01 | Clustering coefficient | 0.00E+00 |
| Neighborhood connectivity | 8.00E+00 | Topological coefficient | 1.00E+00 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| BioCyc | [+] 1 BioCyc Pathways | + | ||||
| 1 | Triacylglycerol degradation | |||||
| KEGG Pathway | [+] 3 KEGG Pathways | + | ||||
| 1 | Glycerolipid metabolism | |||||
| 2 | Metabolic pathways | |||||
| 3 | Retrograde endocannabinoid signaling | |||||
| NetPath Pathway | [+] 1 NetPath Pathways | + | ||||
| 1 | EGFR1 Signaling Pathway | |||||
| Reactome | [+] 2 Reactome Pathways | + | ||||
| 1 | Acyl chain remodeling of DAG and TAG | |||||
| 2 | Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis | |||||
| WikiPathways | [+] 4 WikiPathways | + | ||||
| 1 | Lipid digestion, mobilization, and transport | |||||
| 2 | Glycerophospholipid biosynthesis | |||||
| 3 | GPCR downstream signaling | |||||
| 4 | Effects of PIP2 hydrolysis | |||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 2 | A patent review of Monoacylglycerol Lipase (MAGL) inhibitors (2013-2017).Expert Opin Ther Pat. 2017 Dec;27(12):1341-1351. | |||||
| REF 3 | Selective blockade of 2-arachidonoylglycerol hydrolysis produces cannabinoid behavioral effects. Nat Chem Biol. 2009 Jan;5(1):37-44. | |||||
| REF 4 | Crystal structure of a soluble form of human monoglyceride lipase in complex with an inhibitor at 1.35 ? resolution. Protein Sci. 2011 Apr;20(4):670-83. | |||||
| REF 5 | Design, Synthesis, and Evaluation of Piperazinyl Pyrrolidin-2-ones as a Novel Series of Reversible Monoacylglycerol Lipase Inhibitors. J Med Chem. 2018 Oct 25;61(20):9205-9217. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

