Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T15377
(Former ID: TTDR00513)
|
|||||
| Target Name |
Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 (ATP2A2)
|
|||||
| Synonyms |
SR Ca(2+)Sarcoplasmic/endoplasmic reticulum calcium ATPase 2-ATPase 2; SR Ca(2+)-ATPase 2; SR Ca(2+)-ATPase; SR Ca(2+) pump; SERCA2; Endoplasmic reticulum class 1/2 Ca(2+) ATPase; Cardiacsarcoplasmic reticulum calcium ATPase; CardiacCa2+ ATPase (SERCA2a); Calciumpump 2; Calcium-transporting ATPase sarcoplasmic reticulum type, slow twitch skeletal muscle isoform; Calcium pump 2; ATP2B
Click to Show/Hide
|
|||||
| Gene Name |
ATP2A2
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 2 Target-related Diseases | + | ||||
| 1 | Asthma [ICD-11: CA23] | |||||
| 2 | Heart failure [ICD-11: BD10-BD1Z] | |||||
| Function |
Isoform 2 is involved in the regulation of the contraction/relaxation cycle. Acts as a regulator of TNFSF11-mediated Ca(2+) signaling pathways via its interaction with TMEM64 which is critical for the TNFSF11-induced CREB1 activation and mitochondrial ROS generation necessary for proper osteoclast generation. Association between TMEM64 and SERCA2 in the ER leads to cytosolic Ca (2+) spiking for activation of NFATC1 and production of mitochondrial ROS, thereby triggering Ca (2+) signaling cascades that promote osteoclast differentiation and activation. This magnesium-dependent enzyme catalyzes the hydrolysis of ATP coupled with the translocation of calcium from the cytosol to the sarcoplasmic reticulum lumen.
Click to Show/Hide
|
|||||
| BioChemical Class |
Cation transporting ATPase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 7.2.2.10
|
|||||
| Sequence |
MENAHTKTVEEVLGHFGVNESTGLSLEQVKKLKERWGSNELPAEEGKTLLELVIEQFEDL
LVRILLLAACISFVLAWFEEGEETITAFVEPFVILLILVANAIVGVWQERNAENAIEALK EYEPEMGKVYRQDRKSVQRIKAKDIVPGDIVEIAVGDKVPADIRLTSIKSTTLRVDQSIL TGESVSVIKHTDPVPDPRAVNQDKKNMLFSGTNIAAGKAMGVVVATGVNTEIGKIRDEMV ATEQERTPLQQKLDEFGEQLSKVISLICIAVWIINIGHFNDPVHGGSWIRGAIYYFKIAV ALAVAAIPEGLPAVITTCLALGTRRMAKKNAIVRSLPSVETLGCTSVICSDKTGTLTTNQ MSVCRMFILDRVEGDTCSLNEFTITGSTYAPIGEVHKDDKPVNCHQYDGLVELATICALC NDSALDYNEAKGVYEKVGEATETALTCLVEKMNVFDTELKGLSKIERANACNSVIKQLMK KEFTLEFSRDRKSMSVYCTPNKPSRTSMSKMFVKGAPEGVIDRCTHIRVGSTKVPMTSGV KQKIMSVIREWGSGSDTLRCLALATHDNPLRREEMHLEDSANFIKYETNLTFVGCVGMLD PPRIEVASSVKLCRQAGIRVIMITGDNKGTAVAICRRIGIFGQDEDVTSKAFTGREFDEL NPSAQRDACLNARCFARVEPSHKSKIVEFLQSFDEITAMTGDGVNDAPALKKAEIGIAMG SGTAVAKTASEMVLADDNFSTIVAAVEEGRAIYNNMKQFIRYLISSNVGEVVCIFLTAAL GFPEALIPVQLLWVNLVTDGLPATALGFNPPDLDIMNKPPRNPKEPLISGWLFFRYLAIG CYVGAATVGAAAWWFIAADGGPRVSFYQLSHFLQCKEDNPDFEGVDCAIFESPYPMTMAL SVLVTIEMCNALNSLSENQSLLRMPPWENIWLVGSICLSMSLHFLILYVEPLPLIFQITP LNVTQWLMVLKISLPVILMDETLKFVARNYLEPGKECVQPATKSCSFSACTDGISWPFVL LIMPLVIWVYSTDTNFSDMFWS Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| HIT2.0 ID | T62Z4P | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 2 Clinical Trial Drugs | + | ||||
| 1 | Gallopamil | Drug Info | Phase 2 | Asthma | [2] | |
| 2 | Mydicar | Drug Info | Phase 2 | Heart failure | [3] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Inhibitor | [+] 1 Inhibitor drugs | + | ||||
| 1 | Gallopamil | Drug Info | [1] | |||
| Modulator | [+] 1 Modulator drugs | + | ||||
| 1 | Mydicar | Drug Info | [4] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Adenosine triphosphate | Ligand Info | |||||
| Structure Description | Crystal structure of the SERCA2a in the E2.ATP state | PDB:7BT2 | ||||
| Method | X-ray diffraction | Resolution | 3.00 Å | Mutation | No | [5] |
| PDB Sequence |
MENAHTKTVE
10 EVLGHFGVNE20 STGLSLEQVK30 KLKERWGSNE40 LPAEEGKTLL50 ELVIEQFEDL 60 LVRILLLAAC70 ISFVLAWFEE80 GEETITAFVE90 PFVILLILVA100 NAIVGVWQER 110 NAENAIEALK120 EYEPEMGKVY130 RQDRKSVQRI140 KAKDIVPGDI150 VEIAVGDKVP 160 ADIRLTSIKS170 TTLRVDQSIL180 TGESVSVIKH190 TDPVPDPRAV200 NQDKKNMLFS 210 GTNIAAGKAM220 GVVVATGVNT230 EIGKIRDEMV240 ATEQERTPLQ250 QKLDEFGEQL 260 SKVISLICIA270 VWIINIGHFN280 DPVHGGSWIR290 GAIYYFKIAV300 ALAVAAIPEG 310 LPAVITTCLA320 LGTRRMAKKN330 AIVRSLPSVE340 TLGCTSVICS350 DKTGTLTTNQ 360 MSVCRMFILD370 RVEGDTCSLN380 EFTITGSTYA390 PIGEVHKDDK400 PVNCHQYDGL 410 VELATICALC420 NDSALDYNEA430 KGVYEKVGEA440 TETALTCLVE450 KMNVFDTELK 460 GLSKIERANA470 CNSVIKQLMK480 KEFTLEFSRD490 RKSMSVYCTP500 NKPSRTSMSK 510 MFVKGAPEGV520 IDRCTHIRVG530 STKVPMTSGV540 KQKIMSVIRE550 WGSGSDTLRC 560 LALATHDNPL570 RREEMHLEDS580 ANFIKYETNL590 TFVGCVGMLD600 PPRIEVASSV 610 KLCRQAGIRV620 IMITGDNKGT630 AVAICRRIGI640 FGQDEDVTSK650 AFTGREFDEL 660 NPSAQRDACL670 NARCFARVEP680 SHKSKIVEFL690 QSFDEITAMT700 GDGVNDAPAL 710 KKAEIGIAMG720 SGTAVAKTAS730 EMVLADDNFS740 TIVAAVEEGR750 AIYNNMKQFI 760 RYLISSNVGE770 VVCIFLTAAL780 GFPEALIPVQ790 LLWVNLVTDG800 LPATALGFNP 810 PDLDIMNKPP820 RNPKEPLISG830 WLFFRYLAIG840 CYVGAATVGA850 AAWWFIAADG 860 GPRVSFYQLS870 HFLQCKEDNP880 DFEGVDCAIF890 ESPYPMTMAL900 SVLVTIEMCN 910 ALNSLSENQS920 LLRMPPWENI930 WLVGSICLSM940 SLHFLILYVE950 PLPLIFQITP 960 LNVTQWLMVL970 KISLPVILMD980 ETLKFVARNY990 L
|
|||||
|
|
ASP351
3.628
LYS352
3.423
THR353
2.751
GLU439
3.065
THR441
4.542
GLU442
3.040
PHE487
3.283
ARG489
3.220
LYS492
3.472
SER493
4.123
MET494
3.603
LYS514
2.954
GLY515
3.492
|
|||||
| Ligand Name: L-betagamma-meATP | Ligand Info | |||||
| Structure Description | CryoEM structure of SERCA2b T1032stop in E1-2Ca2+-AMPPCP (class1) | PDB:6LN5 | ||||
| Method | Electron microscopy | Resolution | 2.80 Å | Mutation | No | [6] |
| PDB Sequence |
MENAHTKTVE
10 EVLGHFGVNE20 STGLSLEQVK30 KLKERWGSNE40 LPAEEGKTLL50 ELVIEQFEDL 60 LVRILLLAAC70 ISFVLAWFEE80 GEETITAFVE90 PFVILLILVA100 NAIVGVWQER 110 NAENAIEALK120 EYEPEMGKVY130 RQDRKSVQRI140 KAKDIVPGDI150 VEIAVGDKVP 160 ADIRLTSIKS170 TTLRVDQSIL180 TGESVSVIKH190 TDPVPDPRAV200 NQDKKNMLFS 210 GTNIAAGKAM220 GVVVATGVNT230 EIGKIRDEMV240 ATEQERTPLQ250 QKLDEFGEQL 260 SKVISLICIA270 VWIINIGHFN280 DPVHGGSWIR290 GAIYYFKIAV300 ALAVAAIPEG 310 LPAVITTCLA320 LGTRRMAKKN330 AIVRSLPSVE340 TLGCTSVICS350 DKTGTLTTNQ 360 MSVCRMFILD370 RVEGDTCSLN380 EFTITGSTYA390 PIGEVHKDDK400 PVNCHQYDGL 410 VELATICALC420 NDSALDYNEA430 KGVYEKVGEA440 TETALTCLVE450 KMNVFDTELK 460 GLSKIERANA470 CNSVIKQLMK480 KEFTLEFSRD490 RKSMSVYCTP500 NKPSRTSMSK 510 MFVKGAPEGV520 IDRCTHIRVG530 STKVPMTSGV540 KQKIMSVIRE550 WGSGSDTLRC 560 LALATHDNPL570 RREEMHLEDS580 ANFIKYETNL590 TFVGCVGMLD600 PPRIEVASSV 610 KLCRQAGIRV620 IMITGDNKGT630 AVAICRRIGI640 FGQDEDVTSK650 AFTGREFDEL 660 NPSAQRDACL670 NARCFARVEP680 SHKSKIVEFL690 QSFDEITAMT700 GDGVNDAPAL 710 KKAEIGIAMG720 SGTAVAKTAS730 EMVLADDNFS740 TIVAAVEEGR750 AIYNNMKQFI 760 RYLISSNVGE770 VVCIFLTAAL780 GFPEALIPVQ790 LLWVNLVTDG800 LPATALGFNP 810 PDLDIMNKPP820 RNPKEPLISG830 WLFFRYLAIG840 CYVGAATVGA850 AAWWFIAADG 860 GPRVSFYQLS870 HFLQCKEDNP880 DFEGVDCAIF890 ESPYPMTMAL900 SVLVTIEMCN 910 ALNSLSENQS920 LLRMPPWENI930 WLVGSICLSM940 SLHFLILYVE950 PLPLIFQITP 960 LNVTQWLMVL970 KISLPVILMD980 ETLKFVARNY990 LEGISWPFVL1020 LIMPLVIWVY 1030 S
|
|||||
|
|
ASP351
2.617
LYS352
3.423
THR353
3.081
THR441
4.436
GLU442
3.139
PHE487
3.244
ARG489
2.865
LYS492
3.730
SER493
4.190
MET494
3.865
LYS514
3.399
GLY515
3.479
ALA516
3.730
PRO517
4.740
|
|||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Calcium signaling pathway | hsa04020 | Affiliated Target |
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| cGMP-PKG signaling pathway | hsa04022 | Affiliated Target |
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| cAMP signaling pathway | hsa04024 | Affiliated Target |
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Cardiac muscle contraction | hsa04260 | Affiliated Target |
|
| Class: Organismal Systems => Circulatory system | Pathway Hierarchy | ||
| Adrenergic signaling in cardiomyocytes | hsa04261 | Affiliated Target |
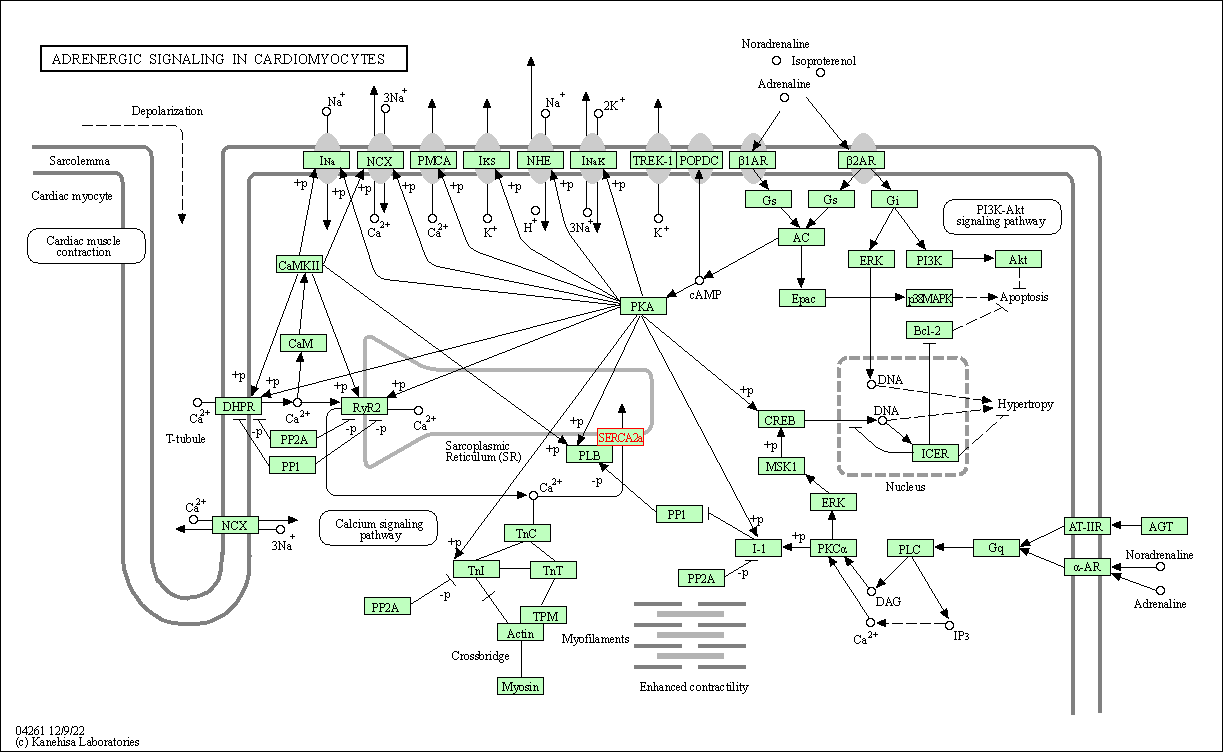
|
| Class: Organismal Systems => Circulatory system | Pathway Hierarchy | ||
| Thyroid hormone signaling pathway | hsa04919 | Affiliated Target |
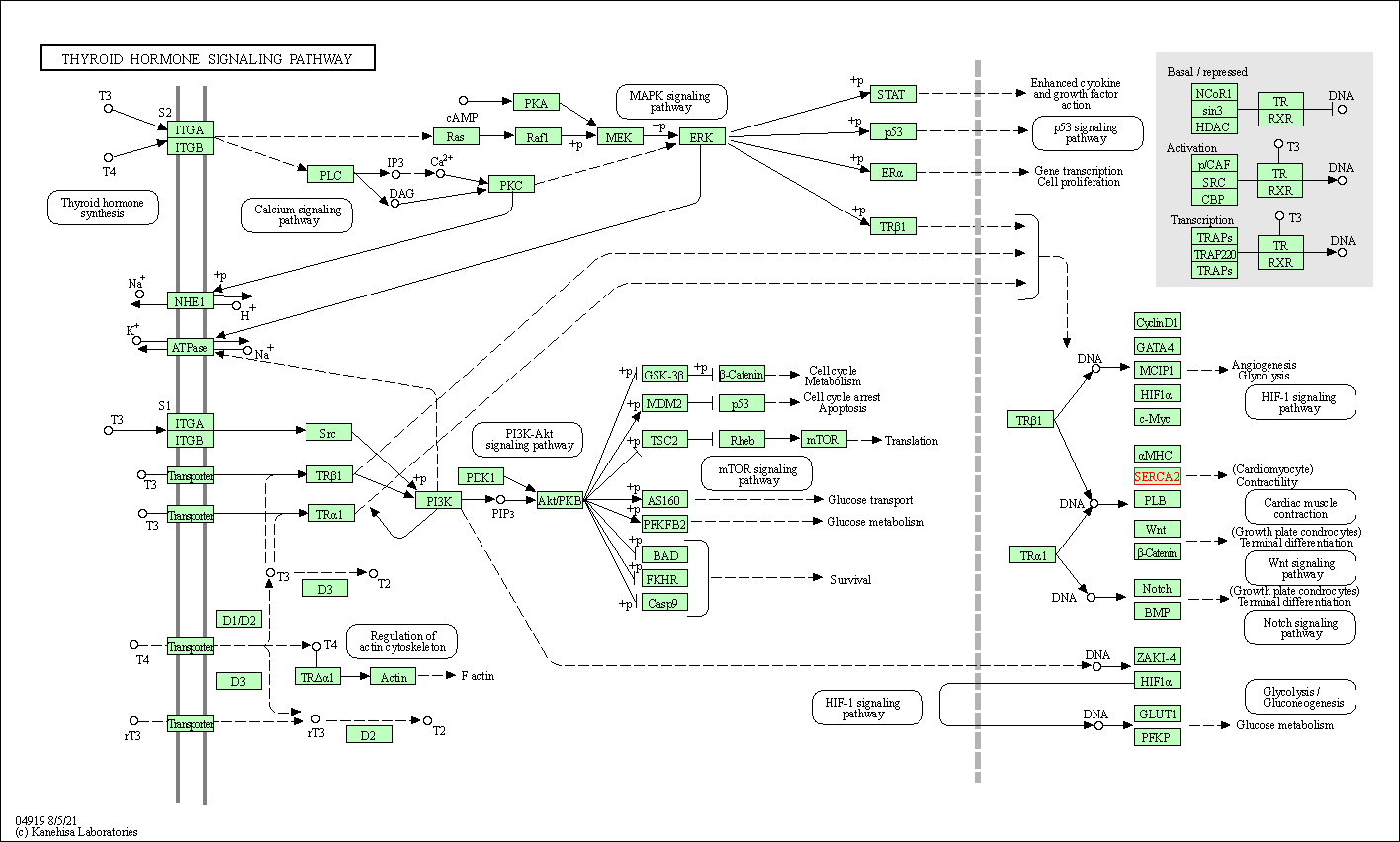
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Pancreatic secretion | hsa04972 | Affiliated Target |
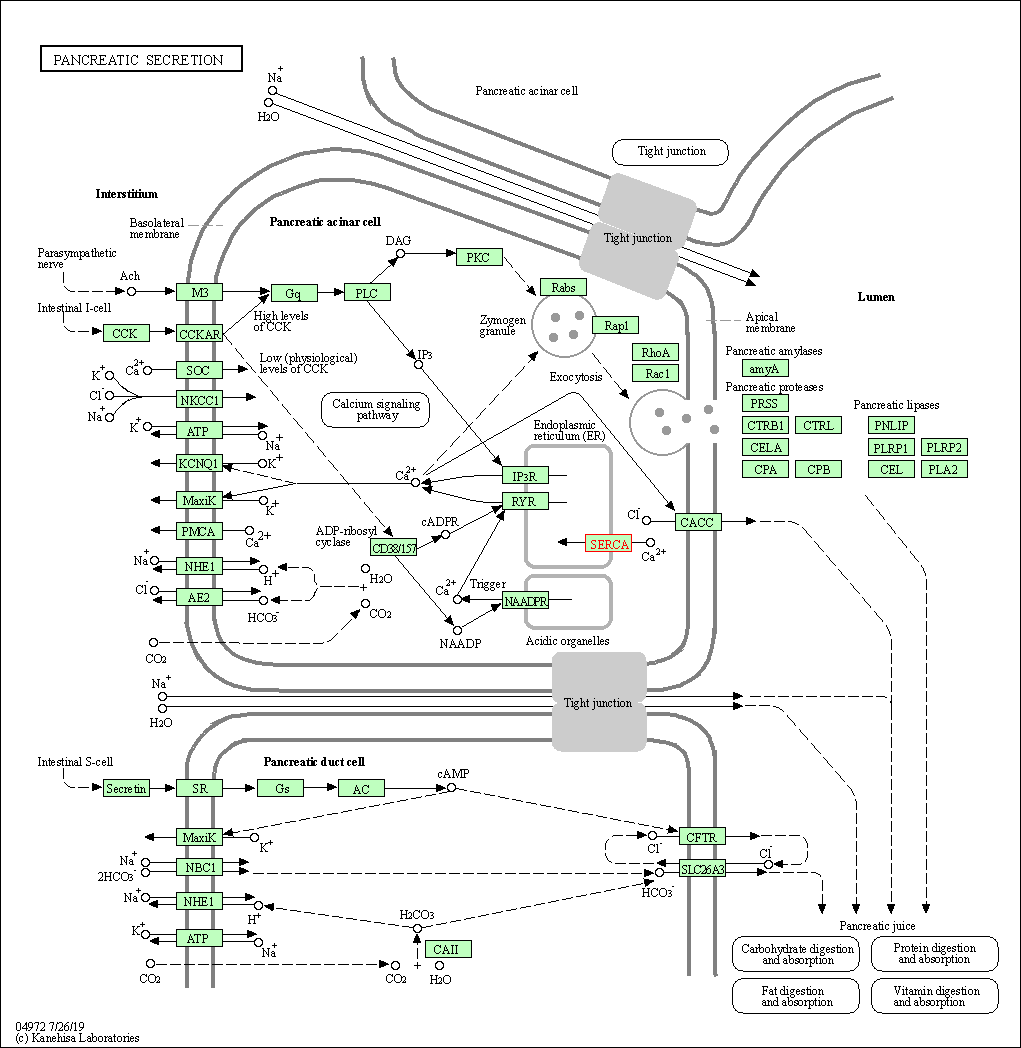
|
| Class: Organismal Systems => Digestive system | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 2 | Degree centrality | 2.15E-04 | Betweenness centrality | 8.09E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 1.25E-01 | Radiality | 1.08E+01 | Clustering coefficient | 0.00E+00 |
| Neighborhood connectivity | 3.00E+00 | Topological coefficient | 5.00E-01 | Eccentricity | 15 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Interaction of D-600 with the transmembrane domain of the sarcoplasmic reticulum Ca(2+)-ATPase. Am J Physiol Cell Physiol. 2000 Jul;279(1):C166-72. | |||||
| REF 2 | ClinicalTrials.gov (NCT00896428) Effects of Gallopamil in Severe Asthma (REMODEL'ASTHME) in University Hospital, Bordeaux. | |||||
| REF 3 | ClinicalTrials.gov (NCT00534703) Investigation of the Safety and Feasibility of AAV1/SERCA2a Gene Transfer in Patients With Chronic Heart Failure and a Left Ventricular Assist Device. U.S. National Institutes of Health. | |||||
| REF 4 | Clinical pipeline report, company report or official report of Celladon. | |||||
| REF 5 | What ATP binding does to the Ca(2+) pump and how nonproductive phosphoryl transfer is prevented in the absence of Ca(2). Proc Natl Acad Sci U S A. 2020 Aug 4;117(31):18448-18458. | |||||
| REF 6 | Cryo-EM structures of SERCA2b reveal the mechanism of regulation by the luminal extension tail. Sci Adv. 2020 Aug 12;6(33):eabb0147. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

