Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T18477
(Former ID: TTDC00018)
|
|||||
| Target Name |
Heat shock protein 90 alpha (HSP90A)
|
|||||
| Synonyms |
Renal carcinoma antigen NY-REN-38; Lipopolysaccharide-associated protein 2; LPS-associated protein 2; LAP-2; Heat shock protein HSP 90-alpha; Heat shock 86 kDa; HSPCA; HSPC1; HSP90A; HSP86; HSP 86
Click to Show/Hide
|
|||||
| Gene Name |
HSP90AA1
|
|||||
| Target Type |
Successful target
|
[1] | ||||
| Disease | [+] 2 Target-related Diseases | + | ||||
| 1 | Acute upper respiratory infection [ICD-11: CA07] | |||||
| 2 | Asthma [ICD-11: CA23] | |||||
| Function |
Undergoes a functional cycle that is linked to its ATPase activity which is essential for its chaperone activity. This cycle probably induces conformational changes in the client proteins, thereby causing their activation. Interacts dynamically with various co-chaperones that modulate its substrate recognition, ATPase cycle and chaperone function. Engages with a range of client protein classes via its interaction with various co-chaperone proteins or complexes, that act as adapters, simultaneously able to interact with the specific client and the central chaperone itself. Recruitment of ATP and co-chaperone followed by client protein forms a functional chaperone. After the completion of the chaperoning process, properly folded client protein and co-chaperone leave HSP90 in an ADP-bound partially open conformation and finally, ADP is released from HSP90 which acquires an open conformation for the next cycle. Apart from its chaperone activity, it also plays a role in the regulation of the transcription machinery. HSP90 and its co-chaperones modulate transcription at least at three different levels. In the first place, they alter the steady-state levels of certain transcription factors in response to various physiological cues(). Second, they modulate the activity of certain epigenetic modifiers, such as histone deacetylases or DNA methyl transferases, and thereby respond to the change in the environment. Third, they participate in the eviction of histones from the promoter region of certain genes and thereby turn on gene expression. Binds bacterial lipopolysaccharide (LPS) and mediates LPS-induced inflammatory response, including TNF secretion by monocytes. Antagonizes STUB1-mediated inhibition of TGF-beta signaling via inhibition of STUB1-mediated SMAD3 ubiquitination and degradation. Molecular chaperone that promotes the maturation, structural maintenance and proper regulation of specific target proteins involved for instance in cell cycle control and signal transduction.
Click to Show/Hide
|
|||||
| BioChemical Class |
Heat shock protein
|
|||||
| UniProt ID | ||||||
| Sequence |
MPEETQTQDQPMEEEEVETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNSSDALDKIR
YESLTDPSKLDSGKELHINLIPNKQDRTLTIVDTGIGMTKADLINNLGTIAKSGTKAFME ALQAGADISMIGQFGVGFYSAYLVAEKVTVITKHNDDEQYAWESSAGGSFTVRTDTGEPM GRGTKVILHLKEDQTEYLEERRIKEIVKKHSQFIGYPITLFVEKERDKEVSDDEAEEKED KEEEKEKEEKESEDKPEIEDVGSDEEEEKKDGDKKKKKKIKEKYIDQEELNKTKPIWTRN PDDITNEEYGEFYKSLTNDWEDHLAVKHFSVEGQLEFRALLFVPRRAPFDLFENRKKKNN IKLYVRRVFIMDNCEELIPEYLNFIRGVVDSEDLPLNISREMLQQSKILKVIRKNLVKKC LELFTELAEDKENYKKFYEQFSKNIKLGIHEDSQNRKKLSELLRYYTSASGDEMVSLKDY CTRMKENQKHIYYITGETKDQVANSAFVERLRKHGLEVIYMIEPIDEYCVQQLKEFEGKT LVSVTKEGLELPEDEEEKKKQEEKKTKFENLCKIMKDILEKKVEKVVVSNRLVTSPCCIV TSTYGWTANMERIMKAQALRDNSTMGYMAAKKHLEINPDHSIIETLRQKAEADKNDKSVK DLVILLYETALLSSGFSLEDPQTHANRIYRMIKLGLGIDEDDPTADDTSAAVTEEMPPLE GDDDTSRMEEVD Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| ADReCS ID | BADD_A05386 ; BADD_A05675 | |||||
| HIT2.0 ID | T47H3O | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Approved Drug(s) | [+] 2 Approved Drugs | + | ||||
| 1 | Amlexanox | Drug Info | Approved | Respiratory tract inflammation | [2], [3], [4] | |
| 2 | Cromoglicate | Drug Info | Approved | Asthma | [5], [4] | |
| Clinical Trial Drug(s) | [+] 14 Clinical Trial Drugs | + | ||||
| 1 | BIIB-021 | Drug Info | Phase 2 | Breast cancer | [6] | |
| 2 | Efungumab | Drug Info | Phase 2 | Breast cancer | [7] | |
| 3 | KW-2478 | Drug Info | Phase 2 | Solid tumour/cancer | [8] | |
| 4 | NVP-AUY922 | Drug Info | Phase 2 | Multiple myeloma | [9] | |
| 5 | Tanespimycin | Drug Info | Phase 2 | Breast cancer | [10], [11] | |
| 6 | VER 50589 | Drug Info | Phase 2 | Breast cancer | [12] | |
| 7 | SNX-5422 | Drug Info | Phase 1/2 | Haematological malignancy | [13] | |
| 8 | Alvespimycin hydrochloride | Drug Info | Phase 1 | Refractory hematologic malignancy | [14] | |
| 9 | AT13387 | Drug Info | Phase 1 | Melanoma | [15] | |
| 10 | BIIB 028 | Drug Info | Phase 1 | Solid tumour/cancer | [16] | |
| 11 | Debio 0932 | Drug Info | Phase 1 | Solid tumour/cancer | [17] | |
| 12 | PU-AD | Drug Info | Phase 1 | Encephalopathy | [18] | |
| 13 | PU3 | Drug Info | Phase 1 | Solid tumour/cancer | [19] | |
| 14 | RTA-901 | Drug Info | Phase 1 | Diabetic neuropathy | [18] | |
| Discontinued Drug(s) | [+] 4 Discontinued Drugs | + | ||||
| 1 | HBP-347 | Drug Info | Discontinued in Phase 3 | Autoimmune diabetes | [20] | |
| 2 | Geldanamycin | Drug Info | Discontinued in Phase 2 | Peripheral nerve damage | [21] | |
| 3 | IPI-493 | Drug Info | Discontinued in Phase 1 | Gastric adenocarcinoma | [22] | |
| 4 | EC-154 | Drug Info | Terminated | Solid tumour/cancer | [25] | |
| Preclinical Drug(s) | [+] 2 Preclinical Drugs | + | ||||
| 1 | CCT-018159 | Drug Info | Preclinical | Solid tumour/cancer | [23] | |
| 2 | KOS-2484 | Drug Info | Preclinical | Solid tumour/cancer | [24] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Inhibitor | [+] 36 Inhibitor drugs | + | ||||
| 1 | Amlexanox | Drug Info | [26] | |||
| 2 | Cromoglicate | Drug Info | [26] | |||
| 3 | BIIB-021 | Drug Info | [27] | |||
| 4 | NVP-AUY922 | Drug Info | [12], [30] | |||
| 5 | Tanespimycin | Drug Info | [1], [30], [31], [32] | |||
| 6 | VER 50589 | Drug Info | [30] | |||
| 7 | SNX-5422 | Drug Info | [33], [32] | |||
| 8 | Alvespimycin hydrochloride | Drug Info | [34], [35], [30] | |||
| 9 | AT13387 | Drug Info | [36] | |||
| 10 | BIIB 028 | Drug Info | [37] | |||
| 11 | Debio 0932 | Drug Info | [17] | |||
| 12 | PU-AD | Drug Info | [18] | |||
| 13 | PU3 | Drug Info | [30] | |||
| 14 | Geldanamycin | Drug Info | [39], [40], [41] | |||
| 15 | IPI-493 | Drug Info | [42], [43] | |||
| 16 | CCT-018159 | Drug Info | [30] | |||
| 17 | KOS-2484 | Drug Info | [44] | |||
| 18 | EC-154 | Drug Info | [45] | |||
| 19 | 17-desmethoxy-17-aminogeldanamycin | Drug Info | [46] | |||
| 20 | 2-(1H-pyrrol-1-ylcarbonyl)benzene-1,3,5-triol | Drug Info | [47] | |||
| 21 | 2-Methyl-2,4-Pentanediol | Drug Info | [48] | |||
| 22 | 4-(2-methoxyethoxy)-6-methylpyrimidin-2-amine | Drug Info | [47] | |||
| 23 | 6-(3-BROMO-2-NAPHTHYL)-1,3,5-TRIAZINE-2,4-DIAMINE | Drug Info | [47] | |||
| 24 | 8-BENZO[1,3]DIOXOL-,5-YLMETHYL-9-BUTYL-9H- | Drug Info | [47] | |||
| 25 | 9-Butyl-8-(3-Methoxybenzyl)-9h-Purin-6-Amine | Drug Info | [47] | |||
| 26 | 9-Butyl-8-(4-Methoxybenzyl)-9h-Purin-6-Amine | Drug Info | [47] | |||
| 27 | Geldanamycin-estradiol hybrid | Drug Info | [49] | |||
| 28 | GNF-PF-67 | Drug Info | [50] | |||
| 29 | KOSN1559 | Drug Info | [30] | |||
| 30 | Macbecin | Drug Info | [30] | |||
| 31 | PU24S | Drug Info | [30] | |||
| 32 | Radicicol | Drug Info | [40], [41] | |||
| 33 | RHEIN | Drug Info | [50] | |||
| 34 | SNX-2112 | Drug Info | [46] | |||
| 35 | VER-49009 | Drug Info | [51] | |||
| 36 | ZEARALANONE | Drug Info | [50] | |||
| Modulator | [+] 3 Modulator drugs | + | ||||
| 1 | KW-2478 | Drug Info | [29] | |||
| 2 | RTA-901 | Drug Info | [18] | |||
| 3 | HBP-347 | Drug Info | [38] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Adenine | Ligand Info | |||||
| Structure Description | Room-temperature Human Hsp90a-NTD bound to adenine | PDB:7S95 | ||||
| Method | X-ray diffraction | Resolution | 1.71 Å | Mutation | No | [52] |
| PDB Sequence |
EVETFAFQAE
25 IAQLMSLIIN35 TFYSNKEIFL45 RELISNSSDA55 LDKIRYESLT65 DPSKLDSGKE 75 LHINLIPNKQ85 DRTLTIVDTG95 IGMTKADLIN105 NLGTIAKSGT115 KAFMEALQAG 125 ADISMIGQFG135 VGFYSAYLVA145 EKVTVITKHN155 DDEQYAWESS165 AGGSFTVRTD 175 TGEPMGRGTK185 VILHLKEDQT195 EYLEERRIKE205 IVKKHSQFIG215 YPITLFVEKE 225
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Ligand Name: Adenosine triphosphate | Ligand Info | |||||
| Structure Description | Hsp90 N-terminal domain bound to ATP | PDB:3T0Z | ||||
| Method | X-ray diffraction | Resolution | 2.19 Å | Mutation | No | [53] |
| PDB Sequence |
PMEEEEVETF
20 AFQAEIAQLM30 SLIINTFYSN40 KEIFLRELIS50 NSSDALDKIR60 YESLTDPSKL 70 DSGKELHINL80 IPNKQDRTLT90 IVDTGIGMTK100 ADLINNLGTI110 AKSGTKAFME 120 ALQAGADISM130 IGQFGVGFYS140 AYLVAEKVTV150 ITKHNDDEQY160 AWESSAGGSF 170 TVRTDTGEPM180 GRGTKVILHL190 KEDQTEYLEE200 RRIKEIVKKH210 SQFIGYPITL 220 FVE
|
|||||
|
|
GLU47
3.861
ASN51
2.903
SER52
3.966
ASP54
4.205
ALA55
3.286
LYS58
4.937
ASP93
2.948
ILE96
4.203
GLY97
4.196
MET98
3.505
ASN106
3.710
LEU107
3.794
|
|||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Protein processing in endoplasmic reticulum | hsa04141 | Affiliated Target |
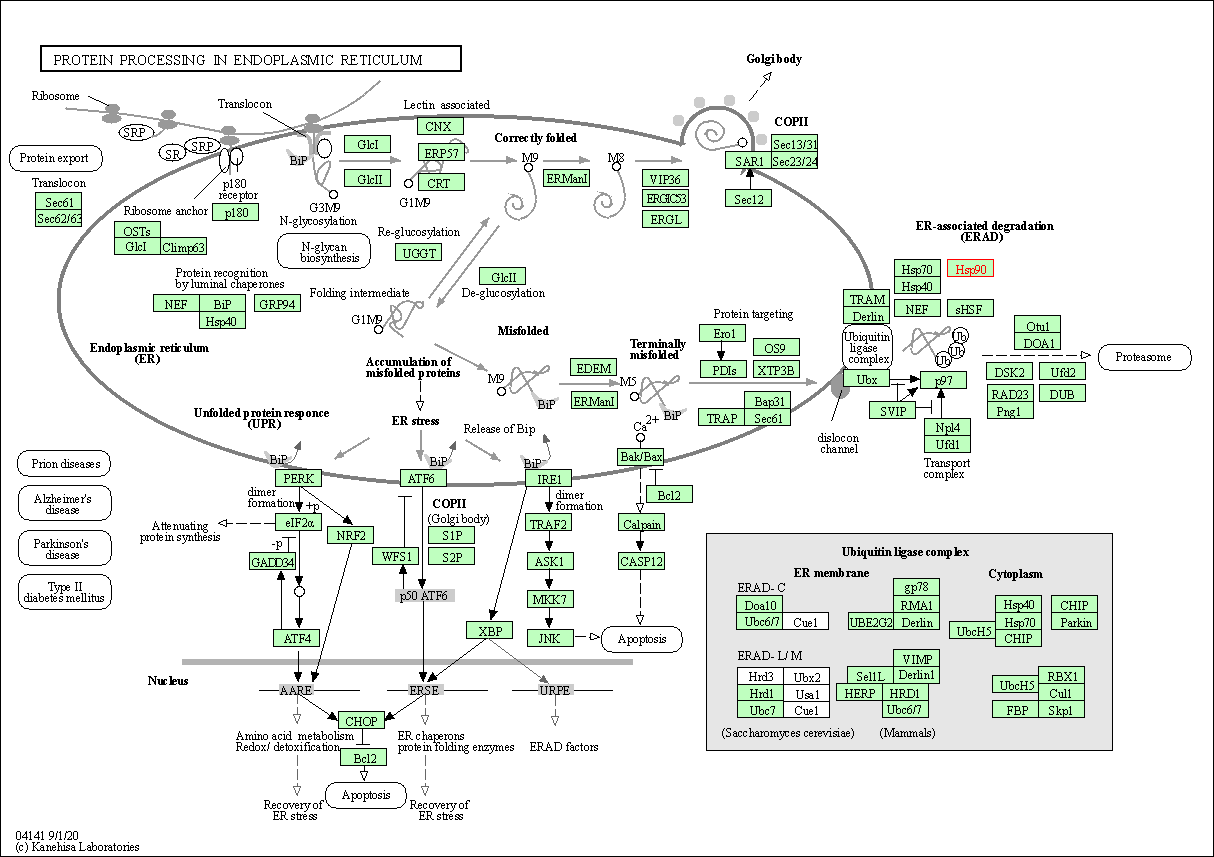
|
| Class: Genetic Information Processing => Folding, sorting and degradation | Pathway Hierarchy | ||
| PI3K-Akt signaling pathway | hsa04151 | Affiliated Target |
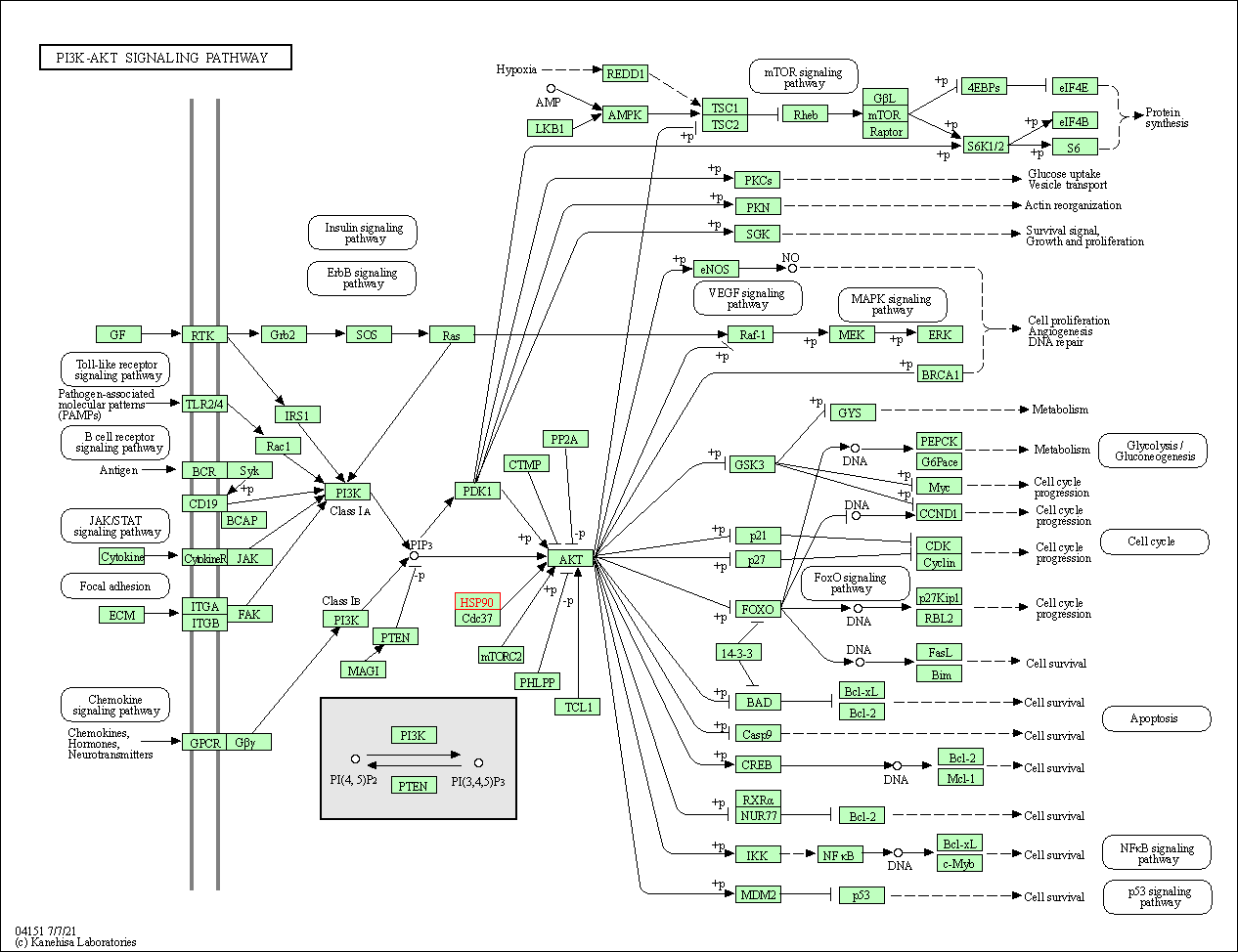
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Necroptosis | hsa04217 | Affiliated Target |
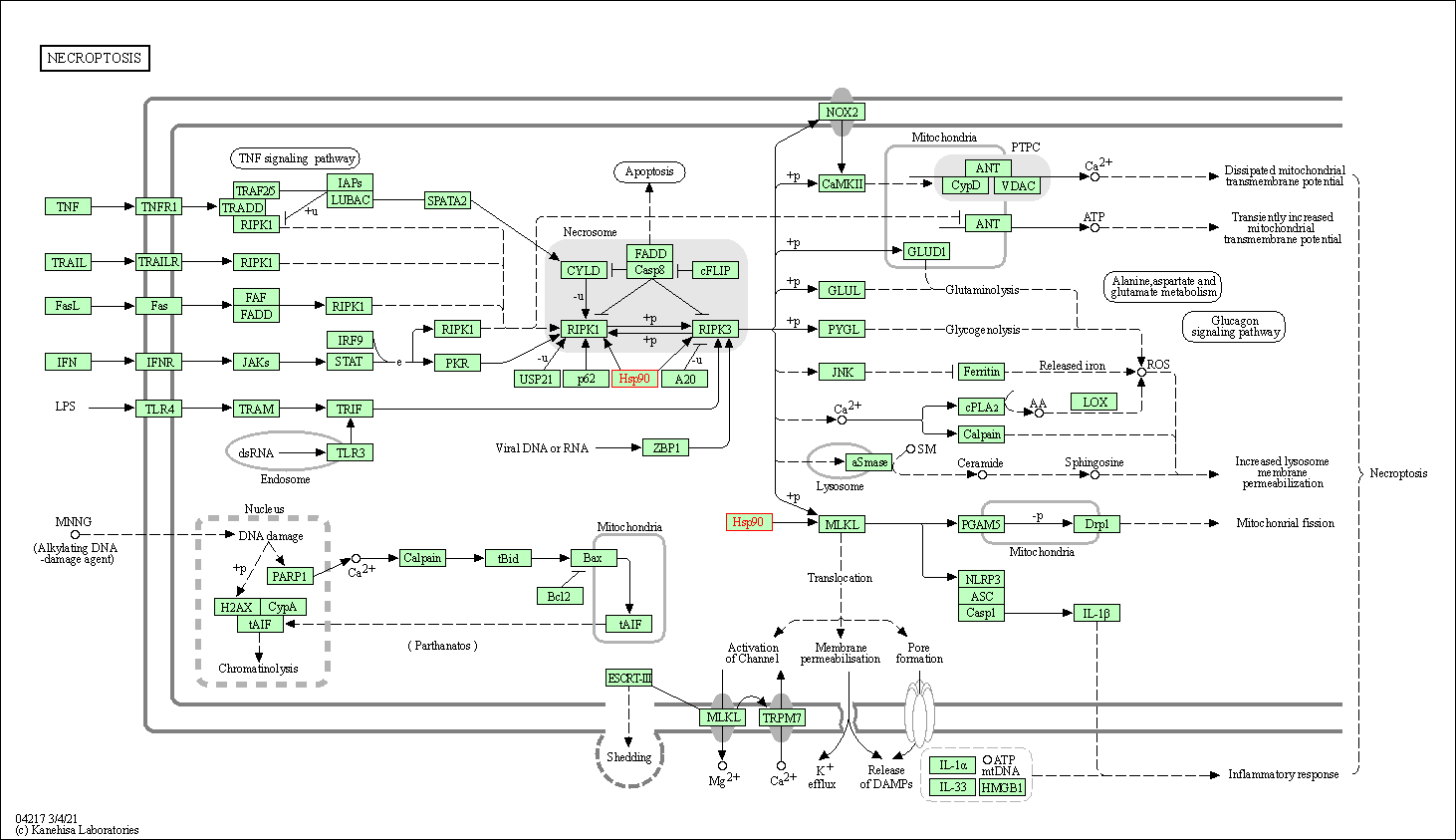
|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Antigen processing and presentation | hsa04612 | Affiliated Target |
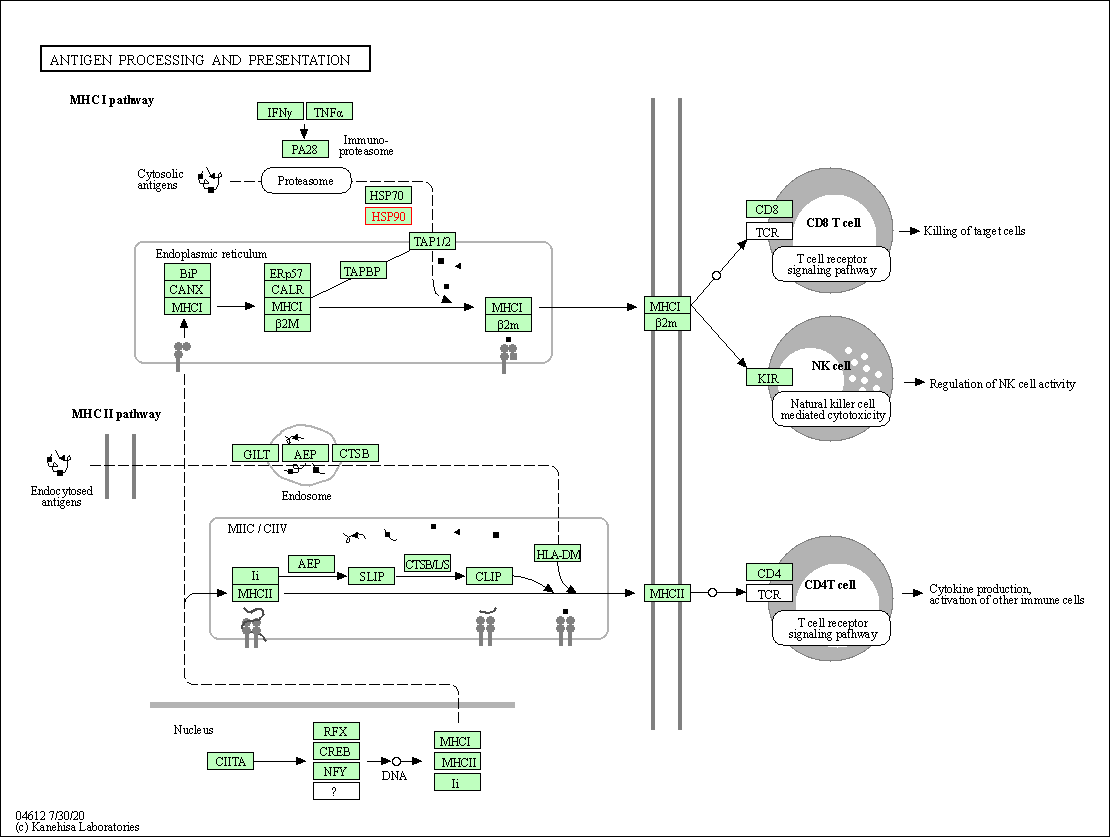
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| NOD-like receptor signaling pathway | hsa04621 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| IL-17 signaling pathway | hsa04657 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Th17 cell differentiation | hsa04659 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Progesterone-mediated oocyte maturation | hsa04914 | Affiliated Target |
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Estrogen signaling pathway | hsa04915 | Affiliated Target |
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 134 | Degree centrality | 1.44E-02 | Betweenness centrality | 4.29E-02 |
|---|---|---|---|---|---|
| Closeness centrality | 2.94E-01 | Radiality | 1.49E+01 | Clustering coefficient | 4.20E-02 |
| Neighborhood connectivity | 3.39E+01 | Topological coefficient | 1.74E-02 | Eccentricity | 10 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) | ||||||
| Drug Resistance Mutation (DRM) | ||||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Tanespimycin: the opportunities and challenges of targeting heat shock protein 90. Expert Opin Investig Drugs. 2009 Jun;18(6):861-8. | |||||
| REF 2 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7113). | |||||
| REF 3 | An inhibitor of the protein kinases TBK1 and IKK- improves obesity-related metabolic dysfunctions in mice. Nat Med. 2013 Mar;19(3):313-21. | |||||
| REF 4 | Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services. 2015 | |||||
| REF 5 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7608). | |||||
| REF 6 | ClinicalTrials.gov (NCT00618319) An Open-Label, 18FDG-PET Pharmacodynamic Assessment of the Effect of BIIB021 in Subjects With Gastrointestinal Stromal Tumors (GIST). U.S. National Institutes of Health. | |||||
| REF 7 | ClinicalTrials.gov (NCT00847678) Efficacy and Safety of Mycograb as Adjunctive Therapy for Cryptococcal Meningitis in Patients With AIDS. U.S. National Institutes of Health. | |||||
| REF 8 | ClinicalTrials.gov (NCT01063907) A Study of KW-2478 in Combination With Bortezomib in Subjects With Relapsed and/or Refractory Multiple Myeloma. U.S. National Institutes of Health. | |||||
| REF 9 | ClinicalTrials.gov (NCT01854034) Phase 2 Study of AUY922 in NSCLC Patients With Exon 20 Insertion Mutations in EGFR. U.S. National Institutes of Health. | |||||
| REF 10 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7751). | |||||
| REF 11 | Emerging therapies for multiple myeloma. Expert Opin Emerg Drugs. 2009 Mar;14(1):99-127. | |||||
| REF 12 | The HSP90 inhibitor NVP-AUY922 potently inhibits non-small cell lung cancer growth. Mol Cancer Ther. 2013 Jun;12(6):890-900. | |||||
| REF 13 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 14 | ClinicalTrials.gov (NCT00089271) 17-DMAG in Treating Patients With Metastatic or Unresectable Solid Tumors or Lymphomas. U.S. National Institutes of Health. | |||||
| REF 15 | Clinical pipeline report, company report or official report of Astex Pharmaceuticals. | |||||
| REF 16 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800020039) | |||||
| REF 17 | Clinical pipeline report, company report or official report of Debiopharm (2011). | |||||
| REF 18 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 19 | Advances in the clinical development of heat shock protein 90 (Hsp90) inhibitors in cancers. Biochim Biophys Acta. 2012 Mar;1823(3):742-55. | |||||
| REF 20 | Heat shock protein 90 (Hsp90). SciBX 4(18). | |||||
| REF 21 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800005345) | |||||
| REF 22 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800026781) | |||||
| REF 23 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800018517) | |||||
| REF 24 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800019408) | |||||
| REF 25 | Inhibition of heat shock protein 90 (Hsp90) modulates macrophage stimulating-1 receptor (MSR1) expression and signaling, and reduces pancreatic tumor growth. Cancer Res May 1, 2009 69; 2835. | |||||
| REF 26 | Hsp90 is a direct target of the anti-allergic drugs disodium cromoglycate and amlexanox. Biochem J. 2003 Sep 1;374(Pt 2):433-41. | |||||
| REF 27 | BIIB021, an orally available, fully synthetic small-molecule inhibitor of the heat shock protein Hsp90. Mol Cancer Ther. 2009 Apr;8(4):921-9. | |||||
| REF 28 | Efungumab: a novel agent in the treatment of invasive candidiasis. Ann Pharmacother. 2009 Nov;43(11):1818-23. | |||||
| REF 29 | Anti-tumor activity against multiple myeloma by combination of KW-2478, an Hsp90 inhibitor, with bortezomib. Blood Cancer J. 2012 Apr;2(4):e68. | |||||
| REF 30 | Recent advances in Hsp90 inhibitors as antitumor agents. Anticancer Agents Med Chem. 2008 Oct;8(7):761-82. | |||||
| REF 31 | Acquired resistance to 17-allylamino-17-demethoxygeldanamycin (17-AAG, tanespimycin) in glioblastoma cells. Cancer Res. 2009 Mar 1;69(5):1966-75. | |||||
| REF 32 | Targeting Hsp90: small-molecule inhibitors and their clinical development. Curr Opin Pharmacol. 2008 Aug;8(4):370-4. | |||||
| REF 33 | SNX-2112, a selective Hsp90 inhibitor, potently inhibits tumor cell growth, angiogenesis, and osteoclastogenesis in multiple myeloma and other hema... Blood. 2009 Jan 22;113(4):846-55. | |||||
| REF 34 | Stage 1 testing and pharmacodynamic evaluation of the HSP90 inhibitor alvespimycin (17-DMAG, KOS-1022) by the pediatric preclinical testing program. Pediatr Blood Cancer. 2008 Jul;51(1):34-41. | |||||
| REF 35 | Anti-proliferative activity of heat shock protein (Hsp) 90 inhibitors via beta-catenin/TCF7L2 pathway in adult T cell leukemia cells. Cancer Lett. 2009 Oct 18;284(1):62-70. | |||||
| REF 36 | Astex Presents Updates on AT13387, its HSP90 Inhibitor, and AT9283, its Multi-Targeted Kinase Inhibitor at the EORTC-NCI-AACR Cancer Conference. Astex. 2008. | |||||
| REF 37 | Phase I study of BIIB028, a selective heat shock protein 90 inhibitor, in patients with refractory metastatic or locally advanced solid tumors. Clin Cancer Res. 2013 Sep 1;19(17):4824-31. | |||||
| REF 38 | US patent application no. 2013,0023,420, Susceptibility to hsp90-inhibitors. | |||||
| REF 39 | Heat shock protein 90 regulates the stability of MEKK3 in HEK293 cells. Cell Immunol. 2009;259(1):49-55. | |||||
| REF 40 | High-throughput screening assay for inhibitors of heat-shock protein 90 ATPase activity. Anal Biochem. 2004 Apr 15;327(2):176-83. | |||||
| REF 41 | Endothelial nitric oxide synthase: the Cinderella of inflammation Trends Pharmacol Sci. 2003 Feb;24(2):91-5. | |||||
| REF 42 | Clinical pipeline report, company report or official report of AstraZeneca (2009). | |||||
| REF 43 | National Cancer Institute. NCI Drug Dictionary. 2009 (CdrID=610131) | |||||
| REF 44 | US patent application no. 2014,0079,636, Targeted therapeutics. | |||||
| REF 45 | WO patent application no. 2013,1734,36, Pre-selection of subjects for therapeutic treatment with an hsp90 inhibitor based on hypoxic status. | |||||
| REF 46 | Heat shock protein 90: inhibitors in clinical trials. J Med Chem. 2010 Jan 14;53(1):3-17. | |||||
| REF 47 | The Protein Data Bank. Nucleic Acids Res. 2000 Jan 1;28(1):235-42. | |||||
| REF 48 | How many drug targets are there Nat Rev Drug Discov. 2006 Dec;5(12):993-6. | |||||
| REF 49 | Synthesis and evaluation of geldanamycin-estradiol hybrids. Bioorg Med Chem Lett. 1999 May 3;9(9):1233-8. | |||||
| REF 50 | In silico identification and biochemical evaluation of novel inhibitors of NRH:quinone oxidoreductase 2 (NQO2). Bioorg Med Chem Lett. 2010 Dec 15;20(24):7331-6. | |||||
| REF 51 | Novel, potent small-molecule inhibitors of the molecular chaperone Hsp90 discovered through structure-based design. J Med Chem. 2005 Jun 30;48(13):4212-5. | |||||
| REF 52 | Water Networks Repopulate Protein-Ligand Interfaces with Temperature. Angew Chem Int Ed Engl. 2022 Aug 1;61(31):e202112919. | |||||
| REF 53 | Structure insights into mechanisms of ATP hydrolysis and the activation of human heat-shock protein 90. Acta Biochim Biophys Sin (Shanghai). 2012 Apr;44(4):300-6. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

