Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T96685
|
|||||
| Target Name |
Hexokinase-2 (HK2)
|
|||||
| Synonyms |
Muscle form hexokinase; Hexokinase type II; HK II
Click to Show/Hide
|
|||||
| Gene Name |
HK2
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Epidermal dysplasias [ICD-11: EK90] | |||||
| Function |
cytosol, membrane, mitochondrial outer membrane, fructokinase activity, glucokinase activity, hexokinase activity, mannokinase activity, apoptotic mitochondrial changes, canonical glycolysis, cellular glucose homeostasis.
Click to Show/Hide
|
|||||
| BioChemical Class |
Hexokinase family
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.7.1.1
|
|||||
| Sequence |
MIASHLLAYFFTELNHDQVQKVDQYLYHMRLSDETLLEISKRFRKEMEKGLGATTHPTAA
VKMLPTFVRSTPDGTEHGEFLALDLGGTNFRVLWVKVTDNGLQKVEMENQIYAIPEDIMR GSGTQLFDHIAECLANFMDKLQIKDKKLPLGFTFSFPCHQTKLDESFLVSWTKGFKSSGV EGRDVVALIRKAIQRRGDFDIDIVAVVNDTVGTMMTCGYDDHNCEIGLIVGTGSNACYME EMRHIDMVEGDEGRMCINMEWGAFGDDGSLNDIRTEFDQEIDMGSLNPGKQLFEKMISGM YMGELVRLILVKMAKEELLFGGKLSPELLNTGRFETKDISDIEGEKDGIRKAREVLMRLG LDPTQEDCVATHRICQIVSTRSASLCAATLAAVLQRIKENKGEERLRSTIGVDGSVYKKH PHFAKRLHKTVRRLVPGCDVRFLRSEDGSGKGAAMVTAVAYRLADQHRARQKTLEHLQLS HDQLLEVKRRMKVEMERGLSKETHASAPVKMLPTYVCATPDGTEKGDFLALDLGGTNFRV LLVRVRNGKWGGVEMHNKIYAIPQEVMHGTGDELFDHIVQCIADFLEYMGMKGVSLPLGF TFSFPCQQNSLDESILLKWTKGFKASGCEGEDVVTLLKEAIHRREEFDLDVVAVVNDTVG TMMTCGFEDPHCEVGLIVGTGSNACYMEEMRNVELVEGEEGRMCVNMEWGAFGDNGCLDD FRTEFDVAVDELSLNPGKQRFEKMISGMYLGEIVRNILIDFTKRGLLFRGRISERLKTRG IFETKFLSQIESDCLALLQVRAILQHLGLESTCDDSIIVKEVCTVVARRAAQLCGAGMAA VVDRIRENRGLDALKVTVGVDGTLYKLHPHFAKVMHETVKDLAPKCDVSFLQSEDGSGKG AALITAVACRIREAGQR Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| HIT2.0 ID | T71GRQ | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 1 Clinical Trial Drugs | + | ||||
| 1 | VDA-1102 | Drug Info | Phase 2 | Actinic keratosis | [1] | |
| Mode of Action | [+] 1 Modes of Action | + | ||||
| Modulator | [+] 1 Modulator drugs | + | ||||
| 1 | VDA-1102 | Drug Info | [1] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: 2-Deoxy-2-{[(2e)-3-(3,4-Dichlorophenyl)prop-2-Enoyl]amino}-Alpha-D-Glucopyranose | Ligand Info | |||||
| Structure Description | Crystal Structure of Human Hexokinase 2 with cmpd 1, a C-2-substituted glucosamine | PDB:5HG1 | ||||
| Method | X-ray diffraction | Resolution | 2.76 Å | Mutation | No | [2] |
| PDB Sequence |
DQVQKVDQYL
26 YHMRLSDETL36 LEISKRFRKE46 MEKGLGATTH56 PTAAVKMLPT66 FVRSTPDGTE 76 HGEFLALDLG86 GTNFRVLWVK96 VTDNGLQKVE106 MENQIYGTQL126 FDHIAECLAN 136 FMDKLQIKDK146 KLPLGFTFSF156 PCHQTKLDES166 FLVSGRDVVA187 LIRKAIQIDI 203 VAVVNDTVGT213 MMTCGYDDHN223 CEIGLIVGTG233 SNACYMEEMR243 HIDMVEGDEG 253 RMCINMEWGA263 FGDDGSLNDI273 RTEFDQEIDM283 GSLNPGKQLF293 EKMISGMYMG 303 ELVRLILVKM313 AKEELLFGGK323 LSPELLNTGR333 FETKDISDIE343 GEKDGIRKAR 353 EVLMRLGLDP363 TQEDCVATHR373 ICQIVSTRSA383 SLCAATLAAV393 LQRIKENKGE 403 ERLRSTIGVD413 GSVYKKHPHF423 AKRLHKTVRR433 LVPGCDVRFL443 RSEDGSGKGA 453 AMVTAVAYRL463 ADQHRARQKT473 LEHLQLSHDQ483 LLEVKRRMKV493 EMERGLSKET 503 HASAPVKMLP513 TYVCATPDGT523 EKGDFLALDL533 GGTNFRVLLV543 RVRNGGVEMH 556 NKIYAIPQEV566 MHGTGDELFD576 HIVQCIADFL586 EYMGMKGVSL596 PLGFTFSFPC 606 QQNSLDESIL616 LKWTKGFKAS626 GCEGEDVVTL636 LKEAIHRREE646 FDLDVVAVVN 656 DTVGTMMTCG666 FEDPHCEVGL676 IVGTGSNACY686 MEEMRNVELV696 EGEEGRMCVN 706 MEWGAFGDNG716 CLDDFRTEFD726 VAVDELSLNP736 GKQRFEKMIS746 GMYLGEIVRN 756 ILIDFTKRGL766 LFRGRISERL776 KTRGIFETKF786 LSQIESDCLA796 LLQVRAILQH 806 LGLESTCDDS816 IIVKEVCTVV826 ARRAAQLCGA836 GMAAVVDRIR846 ENRGLDALKV 856 TVGVDGTLYK866 LHPHFAKVMH876 ETVKDLAPKC886 DVSFLQSEDG896 SGKGAALITA 906 VACRIRE
|
|||||
|
|
SER155
4.493
PRO157
3.354
ASN208
2.777
ASP209
2.683
THR210
4.427
ILE229
3.181
GLY233
3.078
SER234
2.771
ASN235
2.418
ASN258
4.744
GLU260
2.234
GLY262
4.554
ASN287
4.623
LYS290
3.305
GLN291
3.067
GLU294
2.279
GLY534
4.979
SER603
3.088
|
|||||
| Ligand Name: 5-[[(2~{r},3~{s},4~{r},5~{r},6~{s})-5-[(3-Bromophenyl)carbonylamino]-3,4,6-Tris(Oxidanyl)oxan-2-Yl]methylsulfamoyl]-2-Methyl-Furan-3-Carboxylic Acid | Ligand Info | |||||
| Structure Description | Crystal Structure of Human Hexokinase 2 with cmpd 30, a 2-amino-6-benzenesulfonamide glucosamine | PDB:5HEX | ||||
| Method | X-ray diffraction | Resolution | 2.73 Å | Mutation | No | [2] |
| PDB Sequence |
DQVQKVDQYL
26 YHMRLSDETL36 LEISKRFRKE46 MEKGLGATTH56 PTAAVKMLPT66 FVRSTPDGTE 76 HGEFLALDLG86 GTNFRVLWVK96 VTDNGLQKVE106 MENQIYAIPE116 DISGTQLFDH 129 IAECLANFMD139 KLQIKDKKLP149 LGFTFSFPCH159 QTKLDESFLV169 GRDVVALIRK 191 AIQRRGDFDI201 DIVAVVNDTV211 GTMMTCGYDD221 HNCEIGLIVG231 TGSNACYMEE 241 MRHIDMVEGD251 EGRMCINMEW261 GAFGDDGSLN271 DIRTEFDQEI281 DMGSLNPGKQ 291 LFEKMISGMY301 MGELVRLILV311 KMAKEELLFG321 GKLSPELLNT331 GRFETKDISD 341 IEGEKDGIRK351 AREVLMRLGL361 DPTQEDCVAT371 HRICQIVSTR381 SASLCAATLA 391 AVLQRIKENK401 GEERLRSTIG411 VDGSVYKKHP421 HFAKRLHKTV431 RRLVPGCDVR 441 FLRSEDGSGK451 GAAMVTAVAY461 RLADQHRARQ471 KTLEHLQLSH481 DQLLEVKRRM 491 KVEMERGLSK501 ETHASAPVKM511 LPTYVCATPD521 GTEKGDFLAL531 DLGGTNFRVL 541 LVRVRNGKWG551 GVEMHNKIYA561 IPQEVMHGTG571 DELFDHIVQC581 IADFLEYMGM 591 KGVSLPLGFT601 FSFPCQQNSL611 DESILLKWTK621 GFKASGCEGE631 DVVTLLKEAI 641 HRREEFDLDV651 VAVVNDTVGT661 MMTCGFEDPH671 CEVGLIVGTG681 SNACYMEEMR 691 NVELVEGEEG701 RMCVNMEWGA711 FGDNGCLDDF721 RTEFDVAVDE731 LSLNPGKQRF 741 EKMISGMYLG751 EIVRNILIDF761 TKRGLLFRGR771 ISERLKTRGI781 FETKFLSQIE 791 SDCLALLQVR801 AILQHLGLES811 TCDDSIIVKE821 VCTVVARRAA831 QLCGAGMAAV 841 VDRIRENRGL851 DALKVTVGVD861 GTLYKLHPHF871 AKVMHETVKD881 LAPKCDVSFL 891 QSEDGSGKGA901 ALITAVACRI911
|
|||||
|
|
LYS62
3.815
SER155
2.799
PHE156
3.464
PRO157
3.589
ASN208
3.050
ASP209
2.593
THR210
3.881
ILE229
3.775
GLY231
3.224
THR232
2.954
GLY233
3.227
SER234
3.278
ASN235
3.250
ASN258
4.744
GLU260
2.681
GLY262
4.863
ALA263
3.988
GLN291
3.350
GLU294
2.591
ASP413
3.746
|
|||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Glycolysis / Gluconeogenesis | hsa00010 | Affiliated Target |
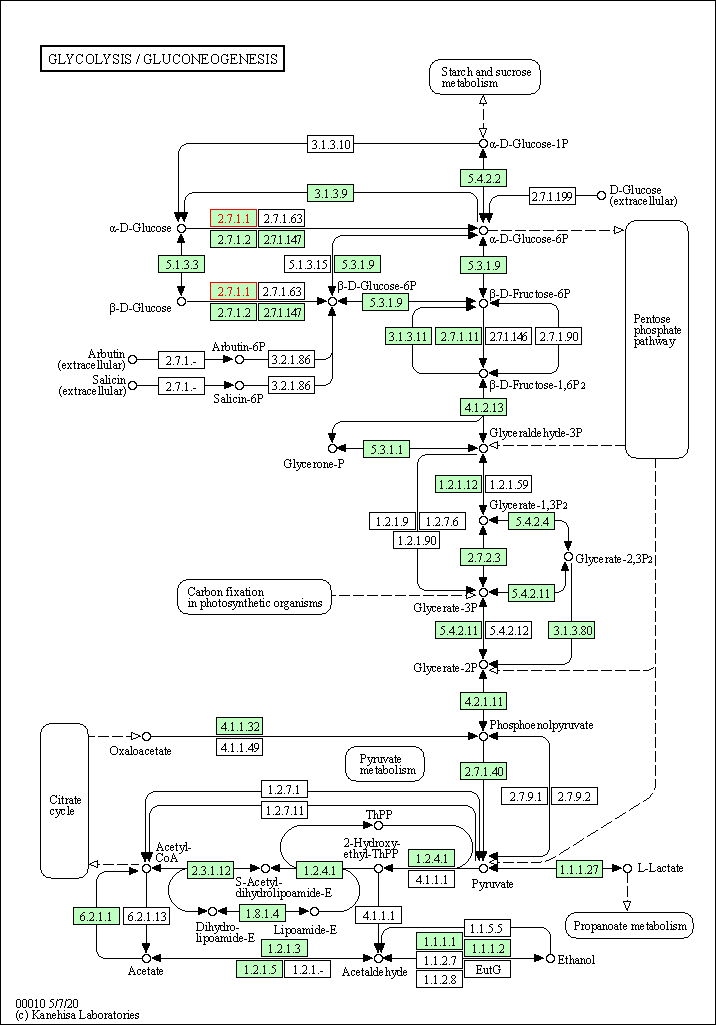
|
| Class: Metabolism => Carbohydrate metabolism | Pathway Hierarchy | ||
| Fructose and mannose metabolism | hsa00051 | Affiliated Target |
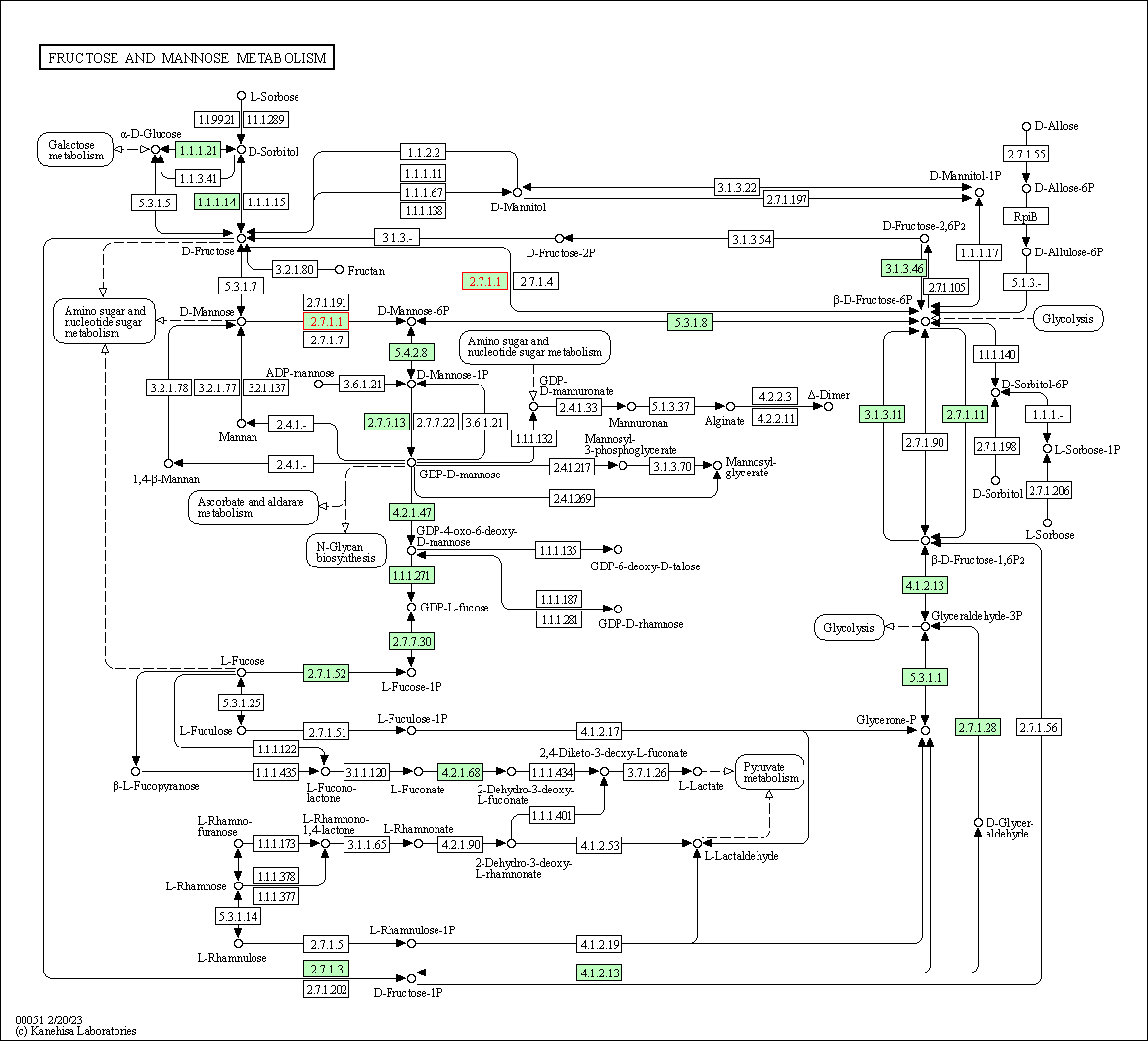
|
| Class: Metabolism => Carbohydrate metabolism | Pathway Hierarchy | ||
| Galactose metabolism | hsa00052 | Affiliated Target |
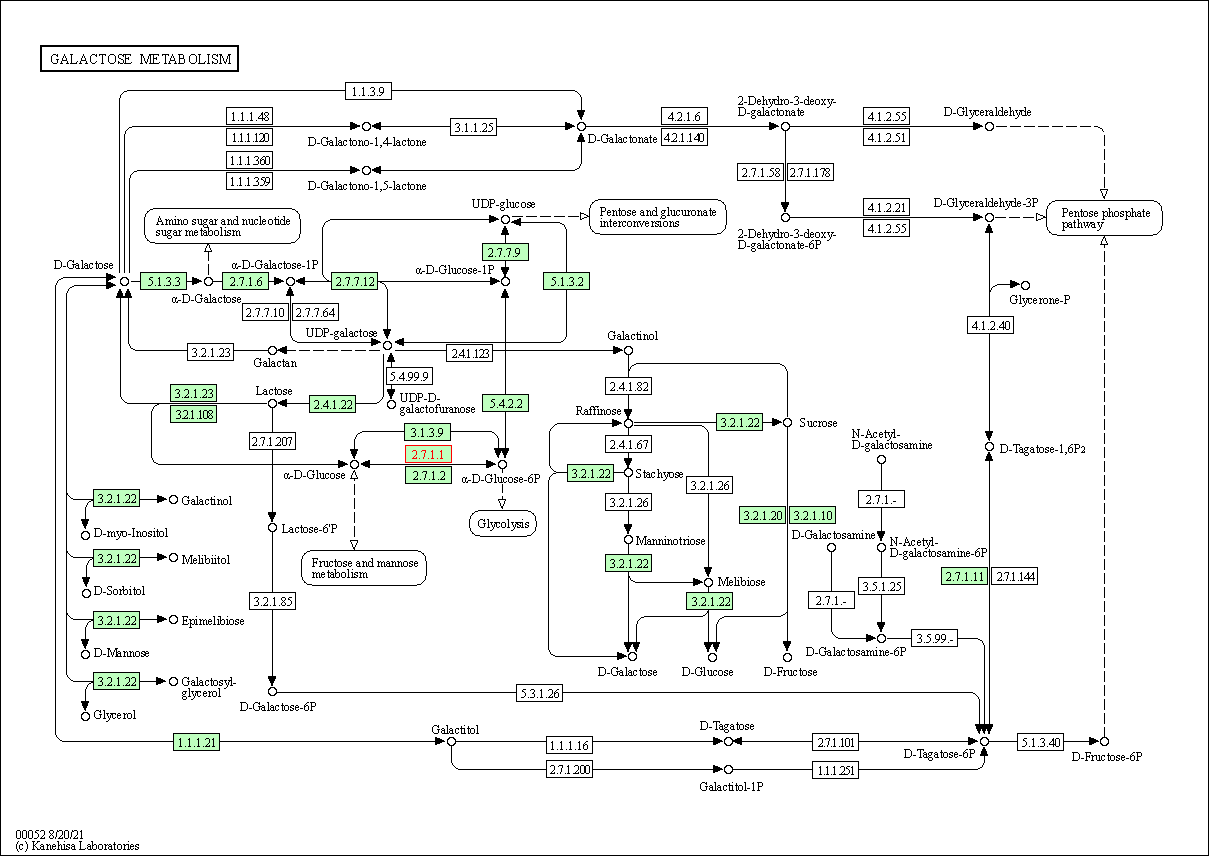
|
| Class: Metabolism => Carbohydrate metabolism | Pathway Hierarchy | ||
| Starch and sucrose metabolism | hsa00500 | Affiliated Target |
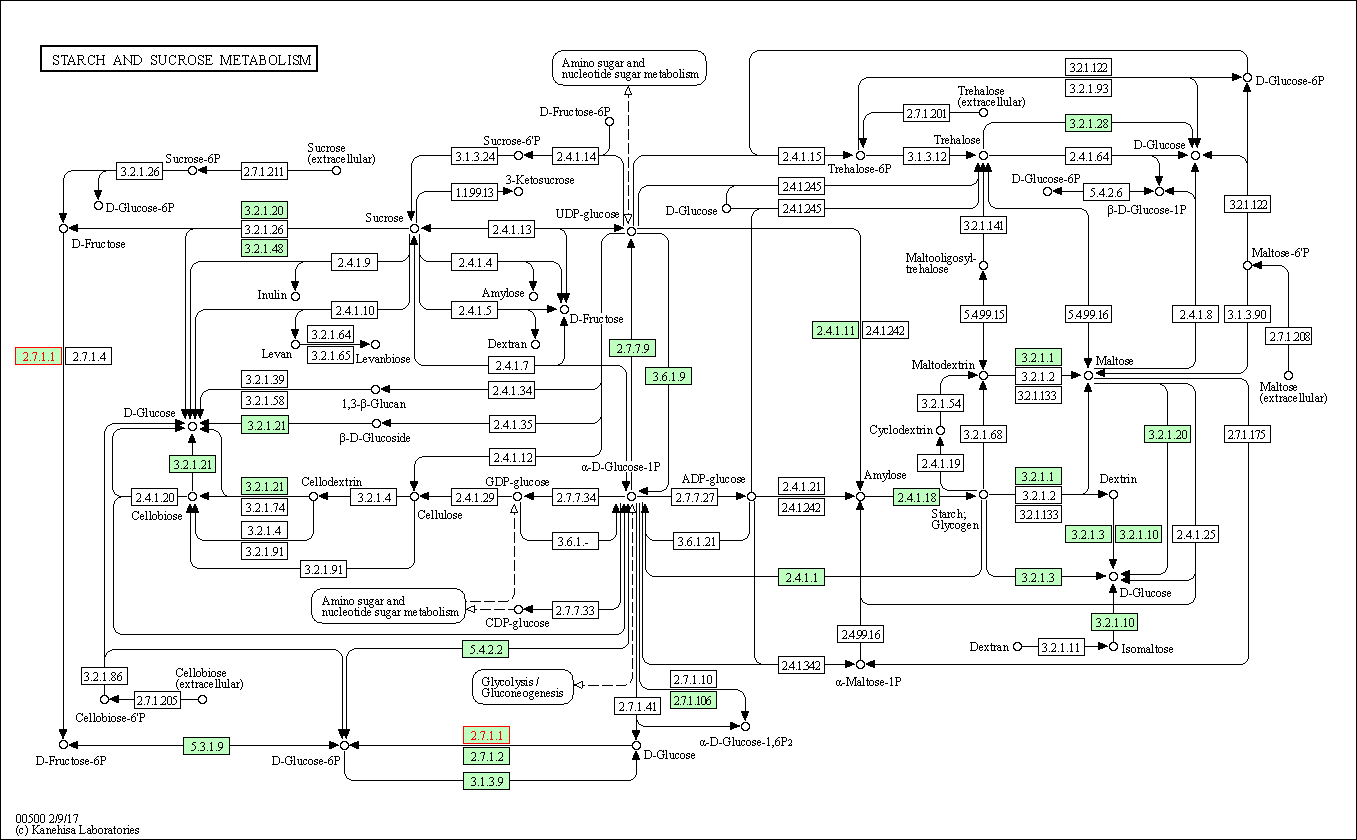
|
| Class: Metabolism => Carbohydrate metabolism | Pathway Hierarchy | ||
| Amino sugar and nucleotide sugar metabolism | hsa00520 | Affiliated Target |
|
| Class: Metabolism => Carbohydrate metabolism | Pathway Hierarchy | ||
| Neomycin, kanamycin and gentamicin biosynthesis | hsa00524 | Affiliated Target |
|
| Class: Metabolism => Biosynthesis of other secondary metabolites | Pathway Hierarchy | ||
| HIF-1 signaling pathway | hsa04066 | Affiliated Target |
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Insulin signaling pathway | hsa04910 | Affiliated Target |
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Carbohydrate digestion and absorption | hsa04973 | Affiliated Target |
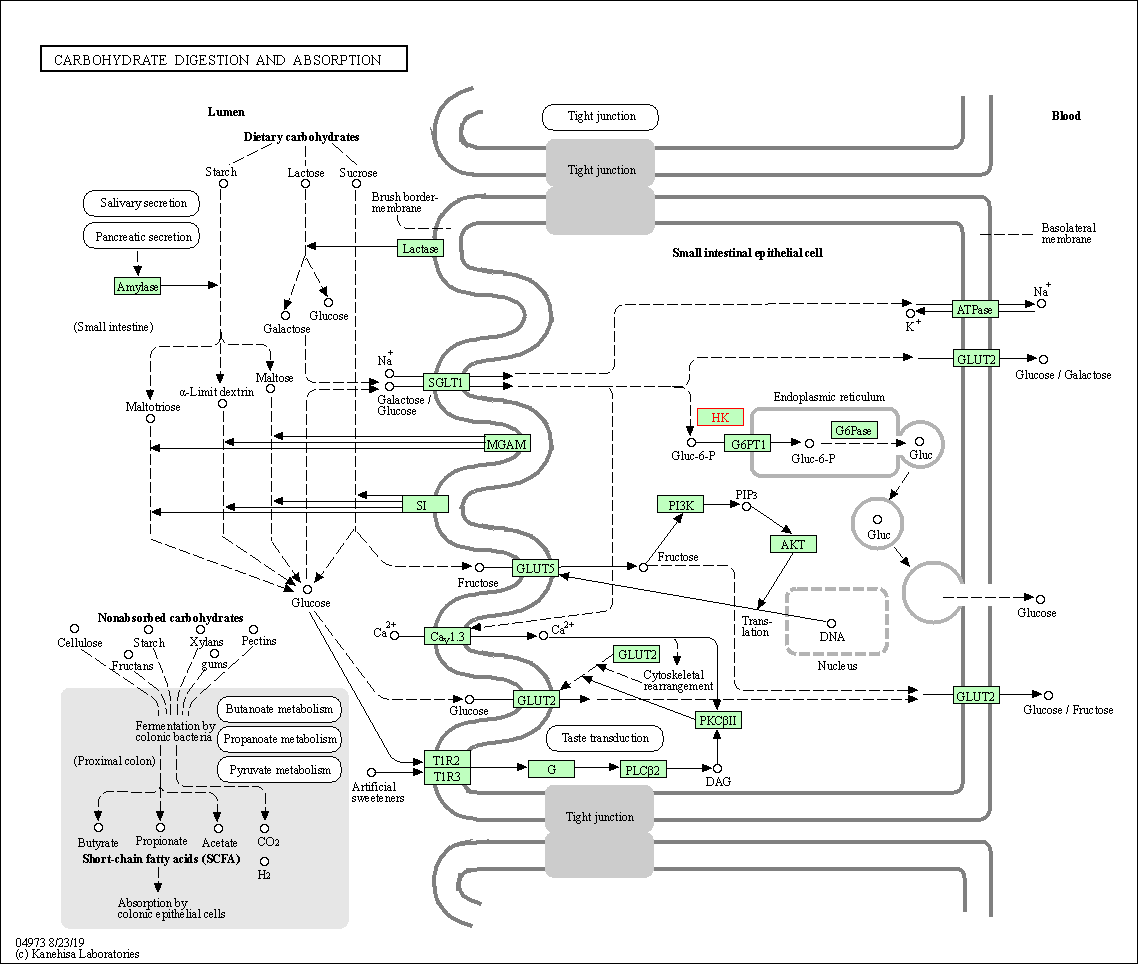
|
| Class: Organismal Systems => Digestive system | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 5 | Degree centrality | 5.37E-04 | Betweenness centrality | 8.75E-05 |
|---|---|---|---|---|---|
| Closeness centrality | 1.97E-01 | Radiality | 1.34E+01 | Clustering coefficient | 2.00E-01 |
| Neighborhood connectivity | 1.90E+01 | Topological coefficient | 2.55E-01 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 13 KEGG Pathways | + | ||||
| 1 | Glycolysis/Gluconeogenesis | |||||
| 2 | Fructose and mannose metabolism | |||||
| 3 | Galactose metabolism | |||||
| 4 | Starch and sucrose metabolism | |||||
| 5 | Amino sugar and nucleotide sugar metabolism | |||||
| 6 | Neomycin, kanamycin and gentamicin biosynthesis | |||||
| 7 | Metabolic pathways | |||||
| 8 | Carbon metabolism | |||||
| 9 | HIF-1 signaling pathway | |||||
| 10 | Insulin signaling pathway | |||||
| 11 | Type II diabetes mellitus | |||||
| 12 | Carbohydrate digestion and absorption | |||||
| 13 | Central carbon metabolism in cancer | |||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 2 | Discovery of a Novel 2,6-Disubstituted Glucosamine Series of Potent and Selective Hexokinase 2 Inhibitors. ACS Med Chem Lett. 2015 Dec 28;7(3):217-22. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

