Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T92557
|
|||||
| Target Name |
Branched-chain-amino-acid transaminase 1 (BCAT1)
|
|||||
| Synonyms |
Protein ECA39; ECA39; Branched-chain-amino-acid aminotransferase, cytosolic; BCT1; BCAT(c)
Click to Show/Hide
|
|||||
| Gene Name |
BCAT1
|
|||||
| Target Type |
Literature-reported target
|
[1] | ||||
| Function |
Catalyzes the first reaction in the catabolism of the essential branched chain amino acids leucine, isoleucine, and valine.
Click to Show/Hide
|
|||||
| BioChemical Class |
Transaminase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.6.1.42
|
|||||
| Sequence |
MKDCSNGCSAECTGEGGSKEVVGTFKAKDLIVTPATILKEKPDPNNLVFGTVFTDHMLTV
EWSSEFGWEKPHIKPLQNLSLHPGSSALHYAVELFEGLKAFRGVDNKIRLFQPNLNMDRM YRSAVRATLPVFDKEELLECIQQLVKLDQEWVPYSTSASLYIRPTFIGTEPSLGVKKPTK ALLFVLLSPVGPYFSSGTFNPVSLWANPKYVRAWKGGTGDCKMGGNYGSSLFAQCEAVDN GCQQVLWLYGEDHQITEVGTMNLFLYWINEDGEEELATPPLDGIILPGVTRRCILDLAHQ WGEFKVSERYLTMDDLTTALEGNRVREMFGSGTACVVCPVSDILYKGETIHIPTMENGPK LASRILSKLTDIQYGREESDWTIVLS Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Gabapentin | Ligand Info | |||||
| Structure Description | Crystal structure of oxidized human cytosolic branched-chain aminotransferase complexed with gabapentin | PDB:2COI | ||||
| Method | X-ray diffraction | Resolution | 1.90 Å | Mutation | Yes | [2] |
| PDB Sequence |
VGTFKAKDLI
31 VTPATILKEK41 PDPNLVFGTV52 FTDHMLTVEW62 SSEFGWEKPH72 IKPLQNLSLH 82 PGSSALHYAV92 ELFEGLKAFR102 GVDNKIRLFQ112 PNLNMDRMYR122 SAVRATLPVF 132 DKEELLECIQ142 QLVKLDQEWV152 PYSTSASLYI162 RPTFIGTEPS172 LGVKKPTKAL 182 LFVLLSPVGP192 YFNPVSLWAN207 PKYVRAWKGG217 TGDCKMGGNY227 GSSLFAQCEA 237 VDNGCQQVLW247 LYGEDHQITE257 VGTMNLFLYW267 INEDGEEELA277 TPPLDGIILP 287 GVTRRCILDL297 AHQWGEFKVS307 ERYLTMDDLT317 TALEGNRVRE327 MFGSGTACVV 337 CPVSDILYKG347 ETIHIPTMEN357 GPKLASRILS367 KLTDIQYGRE377 ERDWTIVL |
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Ligand Name: Pyridoxal phosphate | Ligand Info | |||||
| Structure Description | CRYSTAL STRUCTURE OF HUMAN CYTOSOLIC BRANCHED-CHAIN AMINOTRANSFERASE (BCAT1) IN COMPLEX WITH PLP AND COMPOUND A | PDB:7NWA | ||||
| Method | X-ray diffraction | Resolution | 1.59 Å | Mutation | Yes | [3] |
| PDB Sequence |
VGTFKAKDLI
31 VTPATILKEK41 PDPNNLVFGT51 VFTDHMLTVE61 WSSEFGWEKP71 HIKPLQNLSL 81 HPGSSALHYA91 VELFEGLKAF101 RGVDNKIRLF111 QPNLNMDRMY121 RSAVRATLPV 131 FDKEELLECI141 QQLVKLDQEW151 VPYSTSASLY161 IRPTFIGTEP171 SLGVKKPTKA 181 LLFVLLSPVG191 PYFSSGTFNP201 VSLWANPKYV211 RAWKGGTGDC221 KMGGNYGSSL 231 FAQCEAVDNG241 CQQVLWLYGE251 DHQITEVGTM261 NLFLYWINED271 GEEELATPPL 281 DGIILPGVTR291 RCILDLAHQW301 GEFKVSERYL311 TMDDLTTALE321 GNRVREMFGS 331 GTACVVCPVS341 DILYKGETIH351 IPTMENGPKL361 ASRILSKLTD371 IQYGREERDW 381 TIVLS
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Cysteine and methionine metabolism | hsa00270 | Affiliated Target |
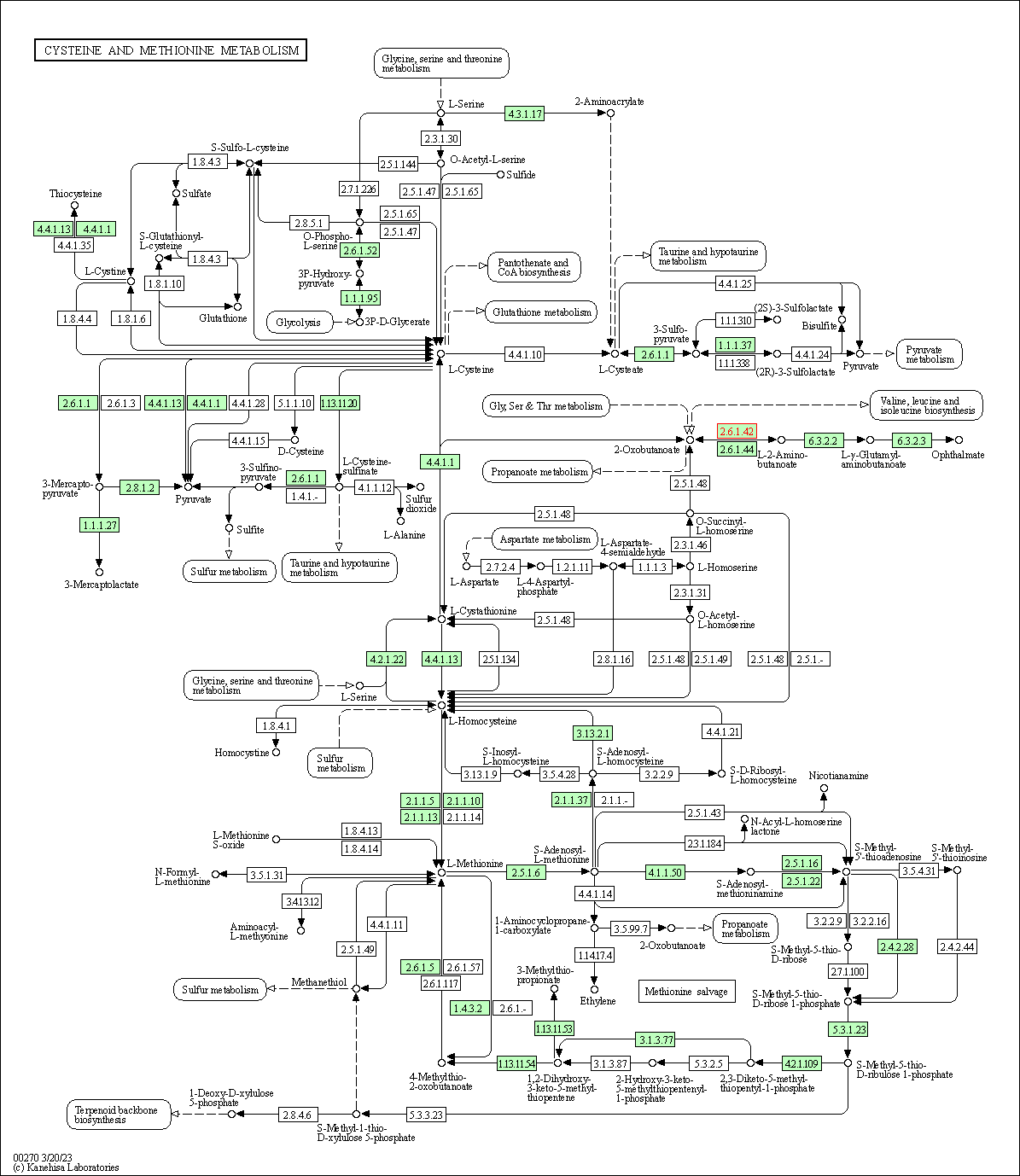
|
| Class: Metabolism => Amino acid metabolism | Pathway Hierarchy | ||
| Valine, leucine and isoleucine degradation | hsa00280 | Affiliated Target |
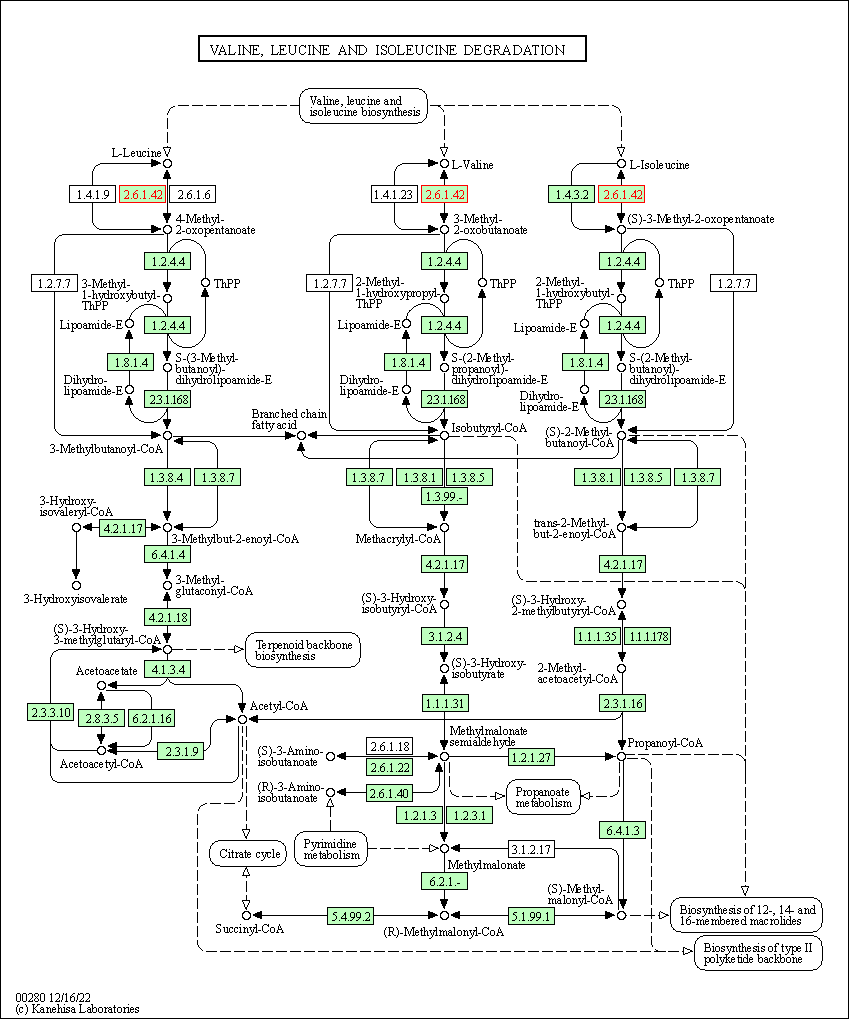
|
| Class: Metabolism => Amino acid metabolism | Pathway Hierarchy | ||
| Valine, leucine and isoleucine biosynthesis | hsa00290 | Affiliated Target |
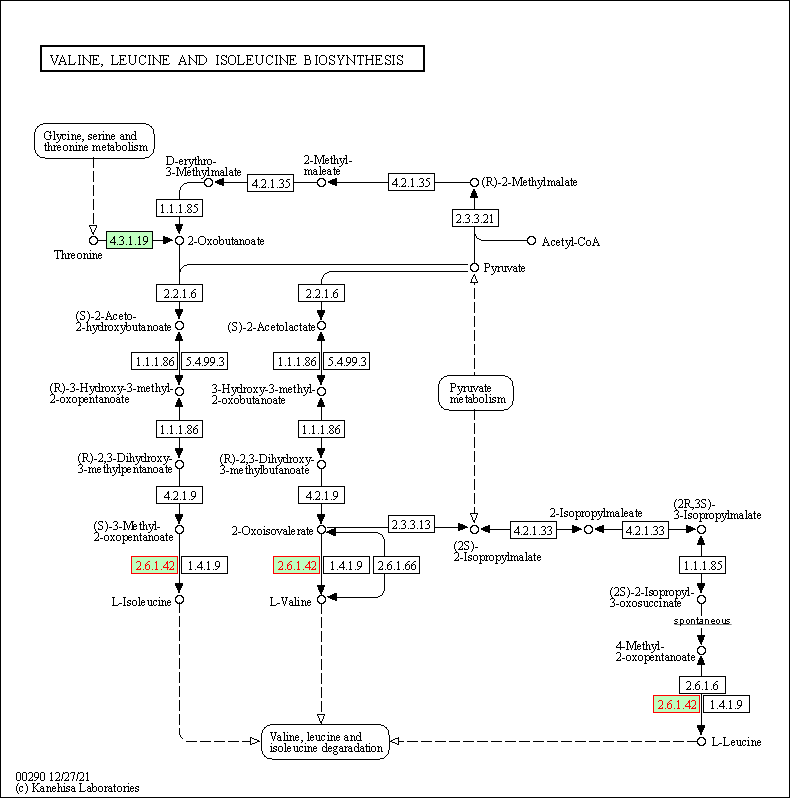
|
| Class: Metabolism => Amino acid metabolism | Pathway Hierarchy | ||
| Pantothenate and CoA biosynthesis | hsa00770 | Affiliated Target |
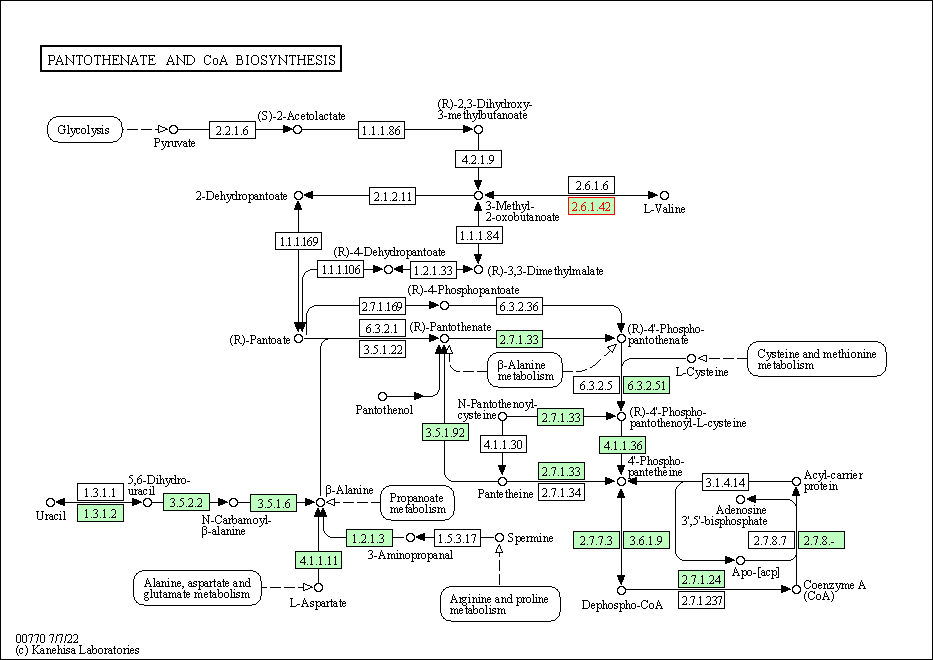
|
| Class: Metabolism => Metabolism of cofactors and vitamins | Pathway Hierarchy | ||
| Degree | 2 | Degree centrality | 2.15E-04 | Betweenness centrality | 0.00E+00 |
|---|---|---|---|---|---|
| Closeness centrality | 1.45E-01 | Radiality | 1.18E+01 | Clustering coefficient | 1.00E+00 |
| Neighborhood connectivity | 1.15E+01 | Topological coefficient | 8.85E-01 | Eccentricity | 14 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 7 KEGG Pathways | + | ||||
| 1 | Cysteine and methionine metabolism | |||||
| 2 | Valine, leucine and isoleucine degradation | |||||
| 3 | Valine, leucine and isoleucine biosynthesis | |||||
| 4 | Pantothenate and CoA biosynthesis | |||||
| 5 | Metabolic pathways | |||||
| 6 | 2-Oxocarboxylic acid metabolism | |||||
| 7 | Biosynthesis of amino acids | |||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Docking and quantitative structure-activity relationship studies for sulfonyl hydrazides as inhibitors of cytosolic human branched-chain amino acid... Mol Divers. 2009 Nov;13(4):493-500. | |||||
| REF 2 | Structural determinants for branched-chain aminotransferase isozyme-specific inhibition by the anticonvulsant drug gabapentin. J Biol Chem. 2005 Nov 4;280(44):37246-56. | |||||
| REF 3 | BAY-069, a Novel (Trifluoromethyl)pyrimidinedione-Based BCAT1/2 Inhibitor and Chemical Probe. J Med Chem. 2022 Nov 10;65(21):14366-14390. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

