Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T83585
(Former ID: TTDI02221)
|
|||||
| Target Name |
Wiskott-Aldrich syndrome protein (WAS)
|
|||||
| Synonyms |
WASp; IMD2
Click to Show/Hide
|
|||||
| Gene Name |
WAS
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Qualitative platelet defect [ICD-11: 3B62] | |||||
| Function |
Important for efficient actin polymerization. Possible regulator of lymphocyte and platelet function. Mediates actin filament reorganization and the formation of actin pedestals upon infection by pathogenic bacteria. In addition to its role in the cytoplasmic cytoskeleton, also promotes actin polymerization in the nucleus, thereby regulating gene transcription and repair of damaged DNA. Promotes homologous recombination (HR) repair in response to DNA damage by promoting nuclear actin polymerization, leading to drive motility of double-strand breaks (DSBs). Effector protein for Rho-type GTPases that regulates actin filament reorganization via its interaction with the Arp2/3 complex.
Click to Show/Hide
|
|||||
| UniProt ID | ||||||
| Sequence |
MSGGPMGGRPGGRGAPAVQQNIPSTLLQDHENQRLFEMLGRKCLTLATAVVQLYLALPPG
AEHWTKEHCGAVCFVKDNPQKSYFIRLYGLQAGRLLWEQELYSQLVYSTPTPFFHTFAGD DCQAGLNFADEDEAQAFRALVQEKIQKRNQRQSGDRRQLPPPPTPANEERRGGLPPLPLH PGGDQGGPPVGPLSLGLATVDIQNPDITSSRYRGLPAPGPSPADKKRSGKKKISKADIGA PSGFKHVSHVGWDPQNGFDVNNLDPDLRSLFSRAGISEAQLTDAETSKLIYDFIEDQGGL EAVRQEMRRQEPLPPPPPPSRGGNQLPRPPIVGGNKGRSGPLPPVPLGIAPPPPTPRGPP PPGRGGPPPPPPPATGRSGPLPPPPPGAGGPPMPPPPPPPPPPPSSGNGPAPPPLPPALV PAGGLAPGGGRGALLDQIRQGIQLNKTPGAPESSALQPPPQSSEGLVGALMHVMQKRSRA IHSSDEGEDQAGDEDEDDEWDD Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 3 Clinical Trial Drugs | + | ||||
| 1 | OTL-103 | Drug Info | Phase 3 | Wiskott-Aldrich syndrome | [2] | |
| 2 | Hematopoietic stem cell gene therapy | Drug Info | Phase 1/2 | Wiskott-Aldrich syndrome | [3] | |
| 3 | WASP gene therapy | Drug Info | Phase 1/2 | Wiskott-Aldrich syndrome | [1] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Replacement | [+] 1 Replacement drugs | + | ||||
| 1 | OTL-103 | Drug Info | [4] | |||
| Modulator | [+] 2 Modulator drugs | + | ||||
| 1 | Hematopoietic stem cell gene therapy | Drug Info | [5] | |||
| 2 | WASP gene therapy | Drug Info | [1], [6] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: (S)-Wiskostatin | Ligand Info | |||||
| Structure Description | Solution structure of the Wiskott-Aldrich Syndrome Protein (WASP) autoinhibited core domain complexed with (S)-wiskostatin, a small molecule inhibitor | PDB:1T84 | ||||
| Method | Solution NMR | Resolution | N.A. | Mutation | No | [7] |
| PDB Sequence |
SGFKHVSHVG
10 WDPQNGFDVN20 NLDPDLRSLF30 SRAGISEAQL40 TDAETSKLIY50 DFIEDQGGLE 60 AVRQEMRRQG70 GSGGSQSSEG80 LVGALMHVMQ90 KRSRAIHSSD100 EGEDQAG |
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
| Protein Name | Pfam ID | Percentage of Identity (%) | E value |
|---|---|---|---|
| Titin (TTN) | 32.530 (27/83) | 2.00E-03 | |
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Chemokine signaling pathway | hsa04062 | Affiliated Target |
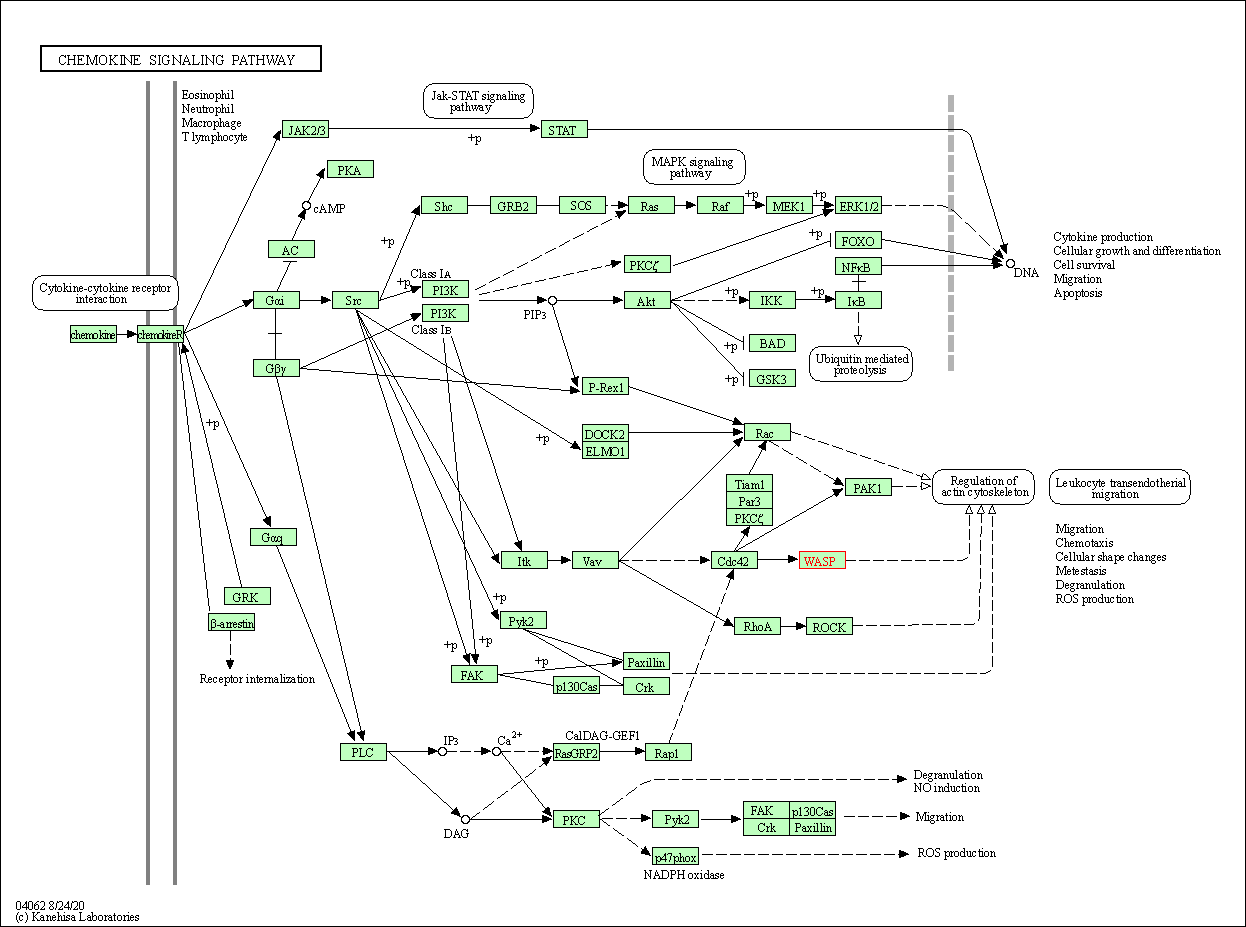
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Adherens junction | hsa04520 | Affiliated Target |
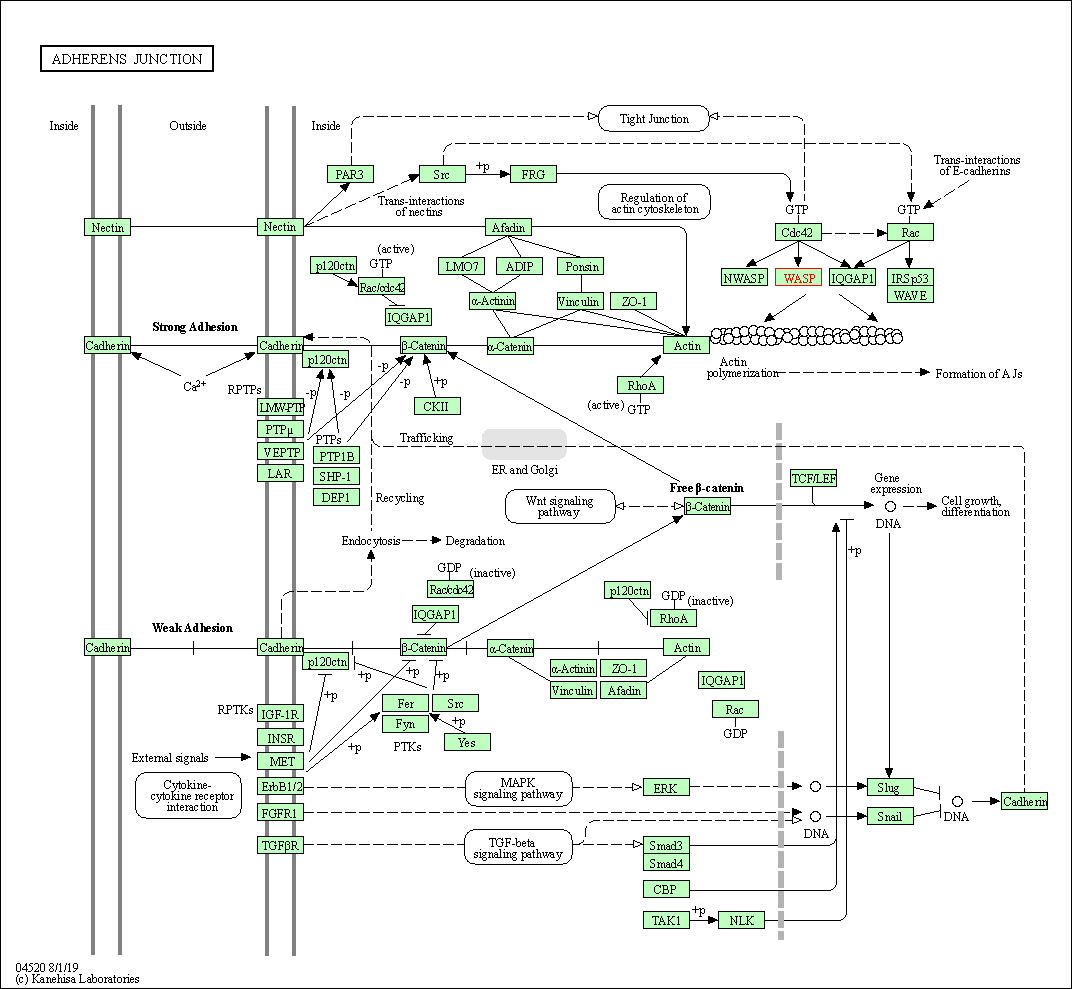
|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
| Tight junction | hsa04530 | Affiliated Target |
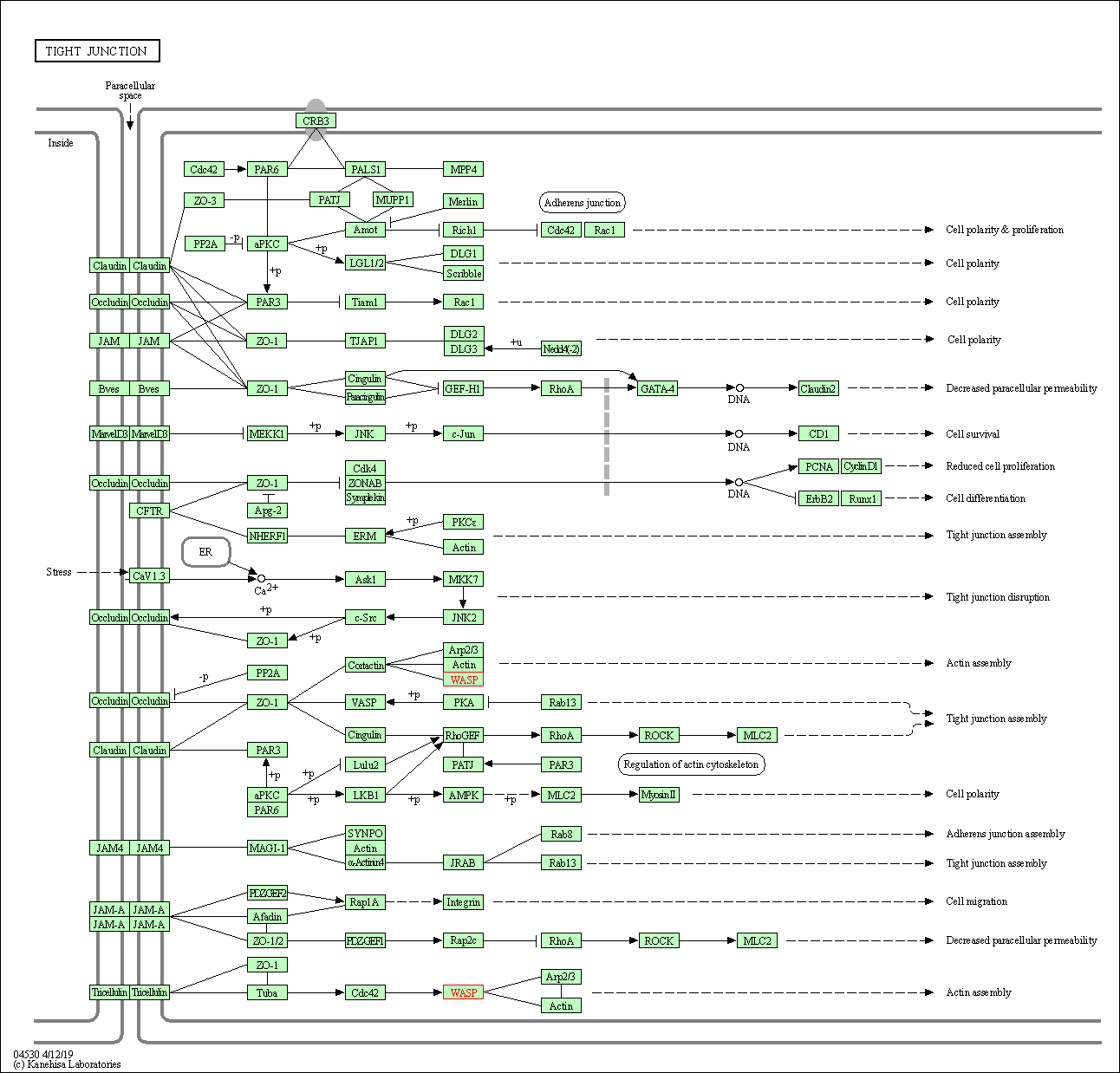
|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
| Fc gamma R-mediated phagocytosis | hsa04666 | Affiliated Target |
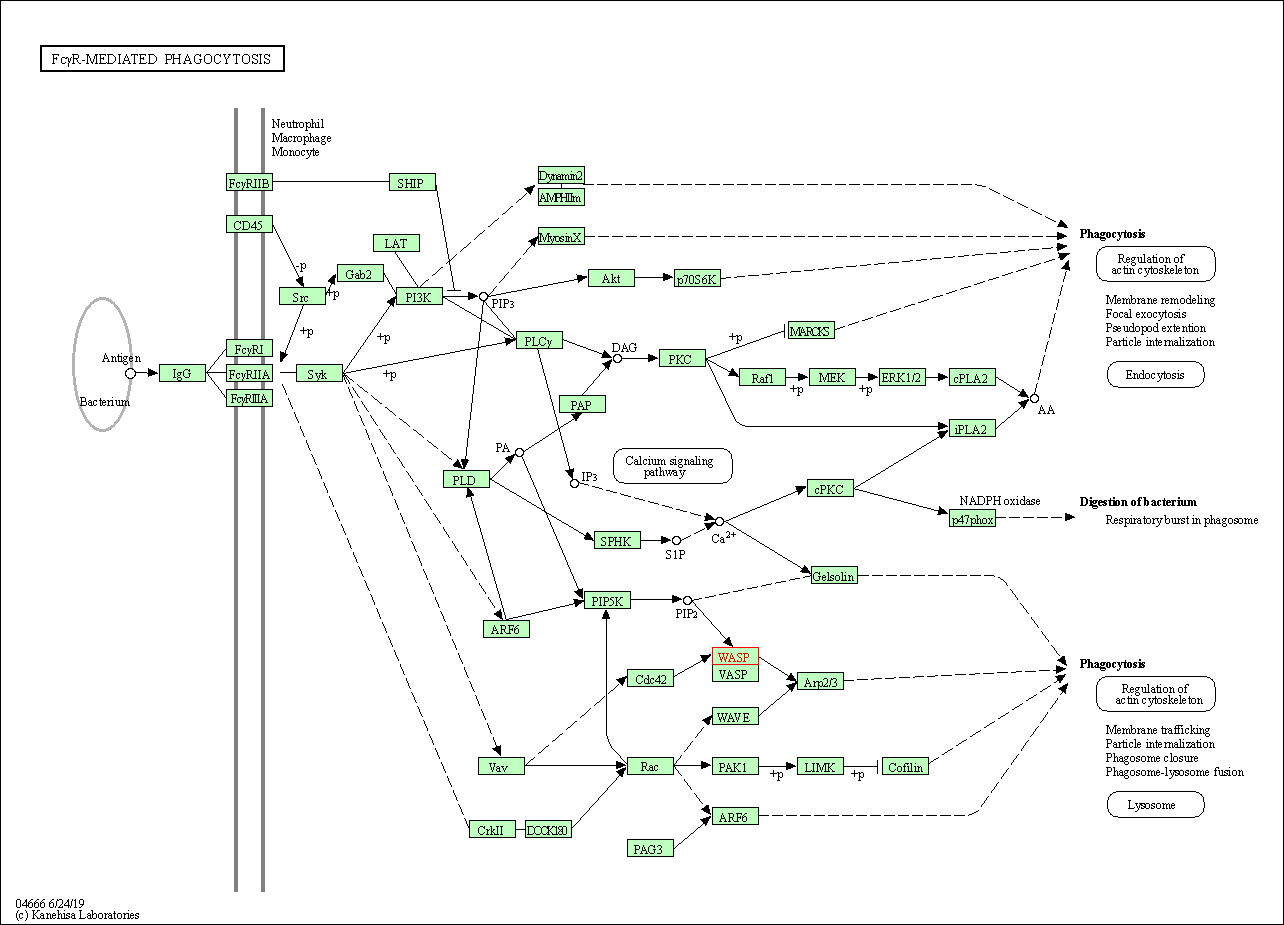
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Degree | 35 | Degree centrality | 3.76E-03 | Betweenness centrality | 1.39E-03 |
|---|---|---|---|---|---|
| Closeness centrality | 2.35E-01 | Radiality | 1.41E+01 | Clustering coefficient | 1.60E-01 |
| Neighborhood connectivity | 3.06E+01 | Topological coefficient | 6.51E-02 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-interacting Proteins | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 9 KEGG Pathways | + | ||||
| 1 | Chemokine signaling pathway | |||||
| 2 | Adherens junction | |||||
| 3 | Fc gamma R-mediated phagocytosis | |||||
| 4 | Regulation of actin cytoskeleton | |||||
| 5 | Bacterial invasion of epithelial cells | |||||
| 6 | Pathogenic Escherichia coli infection | |||||
| 7 | Shigellosis | |||||
| 8 | Salmonella infection | |||||
| 9 | Choline metabolism in cancer | |||||
| Panther Pathway | [+] 1 Panther Pathways | + | ||||
| 1 | T cell activation | |||||
| PID Pathway | [+] 2 PID Pathways | + | ||||
| 1 | TCR signaling in naï | |||||
| 2 | ||||||
| Reactome | [+] 3 Reactome Pathways | + | ||||
| 1 | Generation of second messenger molecules | |||||
| 2 | Regulation of actin dynamics for phagocytic cup formation | |||||
| 3 | RHO GTPases Activate WASPs and WAVEs | |||||
| WikiPathways | [+] 7 WikiPathways | + | ||||
| 1 | TCR Signaling Pathway | |||||
| 2 | G13 Signaling Pathway | |||||
| 3 | Regulation of Actin Cytoskeleton | |||||
| 4 | Human Complement System | |||||
| 5 | Fcgamma receptor (FCGR) dependent phagocytosis | |||||
| 6 | Pathogenic Escherichia coli infection | |||||
| 7 | TCR signaling | |||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | ClinicalTrials.gov (NCT01347242) Gene Therapy for Wiskott-Aldrich Syndrome (WAS). U.S. National Institutes of Health. | |||||
| REF 2 | ClinicalTrials.gov (NCT03837483) A Clinical Study to Evaluate the Use of a Cryopreserved Formulation of OTL-103 in Subjects With Wiskott-Aldrich Syndrome. U.S. National Institutes of Health. | |||||
| REF 3 | ClinicalTrials.gov (NCT02559830) Autologous Hematopoietic Stem Cell Gene Therapy for Metachromatic Leukodystrophy and Adrenoleukodystrophy. | |||||
| REF 4 | Clinical pipeline report, company report or official report of Orchard Therapeutics. | |||||
| REF 5 | Development of hematopoietic stem cell gene therapy for Wiskott-Aldrich syndrome. Curr Opin Mol Ther. 2006 Oct;8(5):390-5. | |||||
| REF 6 | Wiskott-Aldrich syndrome protein-deficient hematopoietic cells can be efficiently mobilized by granulocyte colony-stimulating factor. Haematologica. 2013 August; 98(8): 1300-1308. | |||||
| REF 7 | Chemical inhibition of N-WASP by stabilization of a native autoinhibited conformation. Nat Struct Mol Biol. 2004 Aug;11(8):747-55. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

