Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T82668
(Former ID: TTDI02559)
|
|||||
| Target Name |
Kynurenine oxoglutarate transaminase II (AADAT)
|
|||||
| Synonyms |
Kynurenineoxoglutarate transaminase II; Kynurenineoxoglutarate transaminase 2; Kynurenineoxoglutarate aminotransferase II; Kynurenine/alphaaminoadipate aminotransferase, mitochondrial; Kynurenine aminotransferase II; KAT/AadAT; Alphaaminoadipate aminotransferase; AadAT; 2aminoadipate transaminase; 2aminoadipate aminotransferase
Click to Show/Hide
|
|||||
| Gene Name |
AADAT
|
|||||
| Target Type |
Preclinical target
|
[1] | ||||
| Disease | [+] 2 Target-related Diseases | + | ||||
| 1 | Mild neurocognitive disorder [ICD-11: 6D71] | |||||
| 2 | Schizophrenia [ICD-11: 6A20] | |||||
| Function |
Transaminase with broad substrate specificity. Has transaminase activity towards aminoadipate, kynurenine, methionine and glutamate. Shows activity also towards tryptophan, aspartate and hydroxykynurenine. Accepts a variety of oxo-acids as amino- group acceptors, with a preference for 2-oxoglutarate, 2- oxocaproic acid, phenylpyruvate and alpha-oxo-gamma-methiol butyric acid. Can also use glyoxylate as amino-group acceptor (in vitro).
Click to Show/Hide
|
|||||
| BioChemical Class |
Transaminase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.6.1.39
|
|||||
| Sequence |
MNYARFITAASAARNPSPIRTMTDILSRGPKSMISLAGGLPNPNMFPFKTAVITVENGKT
IQFGEEMMKRALQYSPSAGIPELLSWLKQLQIKLHNPPTIHYPPSQGQMDLCVTSGSQQG LCKVFEMIINPGDNVLLDEPAYSGTLQSLHPLGCNIINVASDESGIVPDSLRDILSRWKP EDAKNPQKNTPKFLYTVPNGNNPTGNSLTSERKKEIYELARKYDFLIIEDDPYYFLQFNK FRVPTFLSMDVDGRVIRADSFSKIISSGLRIGFLTGPKPLIERVILHIQVSTLHPSTFNQ LMISQLLHEWGEEGFMAHVDRVIDFYSNQKDAILAAADKWLTGLAEWHVPAAGMFLWIKV KGINDVKELIEEKAVKMGVLMLPGNAFYVDSSAPSPYLRASFSSASPEQMDVAFQVLAQL IKESL Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Preclinical Drug(s) | [+] 3 Preclinical Drugs | + | ||||
| 1 | (S)-4-(Ethylsulfonyl)benzoylalanine | Drug Info | Preclinical | Cognitive impairment | [2] | |
| 2 | BFF-122 | Drug Info | Preclinical | Cognitive impairment | [2] | |
| 3 | PF-04859989 | Drug Info | Preclinical | Cognitive impairment | [2] | |
| Mode of Action | [+] 1 Modes of Action | + | ||||
| Inhibitor | [+] 10 Inhibitor drugs | + | ||||
| 1 | US10065972, Example 281 | Drug Info | [3] | |||
| 2 | US10065972, Example 396 | Drug Info | [3] | |||
| 3 | US10065972, Example 560 | Drug Info | [3] | |||
| 4 | US8933095, 1 | Drug Info | [4] | |||
| 5 | US8933095, 14 | Drug Info | [5] | |||
| 6 | US8933095, 16 | Drug Info | [4] | |||
| 7 | US8933095, 18 | Drug Info | [5] | |||
| 8 | (S)-4-(Ethylsulfonyl)benzoylalanine | Drug Info | [6] | |||
| 9 | BFF-122 | Drug Info | [2] | |||
| 10 | PF-04859989 | Drug Info | [7] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Pyridoxal phosphate | Ligand Info | |||||
| Structure Description | Crystal structure of human KAT II-inhibitor complex | PDB:2XH1 | ||||
| Method | X-ray diffraction | Resolution | 2.10 Å | Mutation | Yes | [8] |
| PDB Sequence |
ENYARFITAA
10 SAARNPSPIM33 ISLAGGLPNP43 NMFPFKTAVI53 TVENGKTIQF63 GEEMMKRALQ 73 YSPSAGIPEL83 LSWLKQLQIK93 LHNPPTIHYP103 PSQGQMDLCV113 TSGSQQGLCK 123 VFEMIINPGD133 NVLLDEPAYS143 GTLQSLHPLG153 CNIINVASDE163 SGIVPDSLRD 173 ILSRWKPEDA183 KNPQKNTPKF193 LYTVPNGNNP203 TGNSLTSERK213 KEIYELARKY 223 DFLIIEDDPY233 YFLQFNKFRV243 PTFLSMDVDG253 RVIRADSFSK263 IISSGLRIGF 273 LTGPKPLIER283 VILHIQVSTL293 HPSTFNQLMI303 SQLLHEWGEE313 GFMAHVDRVI 323 DFYSNQKDAI333 LAAADKWLTG343 LAEWHVPAAG353 MFLWIKVKGI363 NDVKELIEEK 373 AVKMGVLMLP383 GNAFYVDSSA393 PSPYLRASFS403 SASPEQMDVA413 FQVLAQLIKE 423 SL
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Ligand Name: Pyridoxamine-5'-Phosphate | Ligand Info | |||||
| Structure Description | The crystal structure of human kynurenine aminotransferase II in complex with kynurenine | PDB:2R2N | ||||
| Method | X-ray diffraction | Resolution | 1.95 Å | Mutation | No | [9] |
| PDB Sequence |
MNYARFITAA
10 SAARNPSPIR20 TMTDILSRGP30 KSMISLAGGL40 PNPNMFPFKT50 AVITVENGKT 60 IQFGEEMMKR70 ALQYSPSAGI80 PELLSWLKQL90 QIKLHNPPTI100 HYPPSQGQMD 110 LCVTSGSQQG120 LCKVFEMIIN130 PGDNVLLDEP140 AYSGTLQSLH150 PLGCNIINVA 160 SDESGIVPDS170 LRDILSRWKP180 EDAKNPQKNT190 PKFLYTVPNG200 NNPTGNSLTS 210 ERKKEIYELA220 RKYDFLIIED230 DPYYFLQFNK240 FRVPTFLSMD250 VDGRVIRADS 260 FSKIISSGLR270 IGFLTGPKPL280 IERVILHIQV290 STLHPSTFNQ300 LMISQLLHEW 310 GEEGFMAHVD320 RVIDFYSNQK330 DAILAAADKW340 LTGLAEWHVP350 AAGMFLWIKV 360 KGINDVKELI370 EEKAVKMGVL380 MLPGNAFYVD390 SSAPSPYLRA400 SFSSASPEQM 410 DVAFQVLAQL420 IKESL
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Lysine degradation | hsa00310 | Affiliated Target |
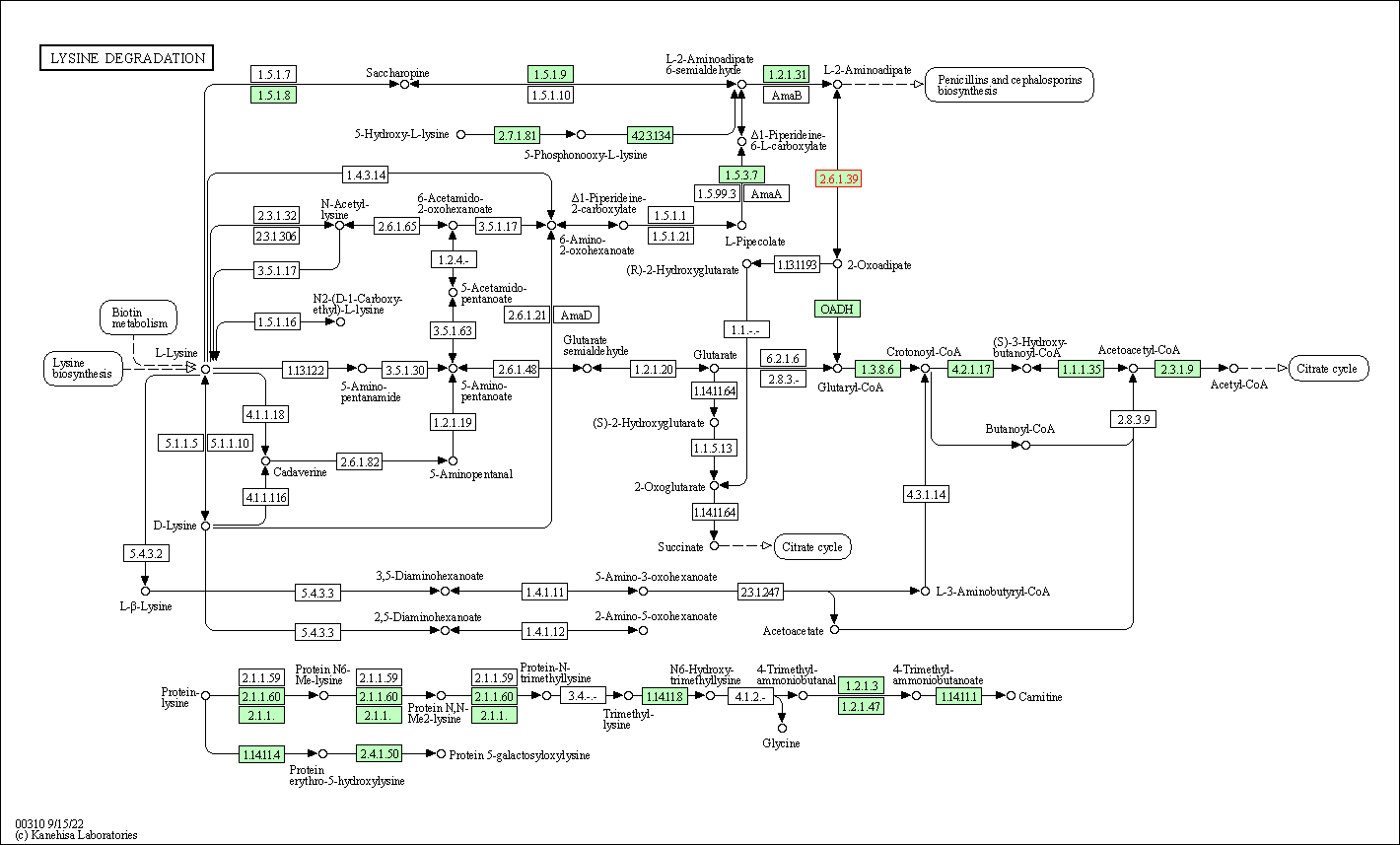
|
| Class: Metabolism => Amino acid metabolism | Pathway Hierarchy | ||
| Tryptophan metabolism | hsa00380 | Affiliated Target |
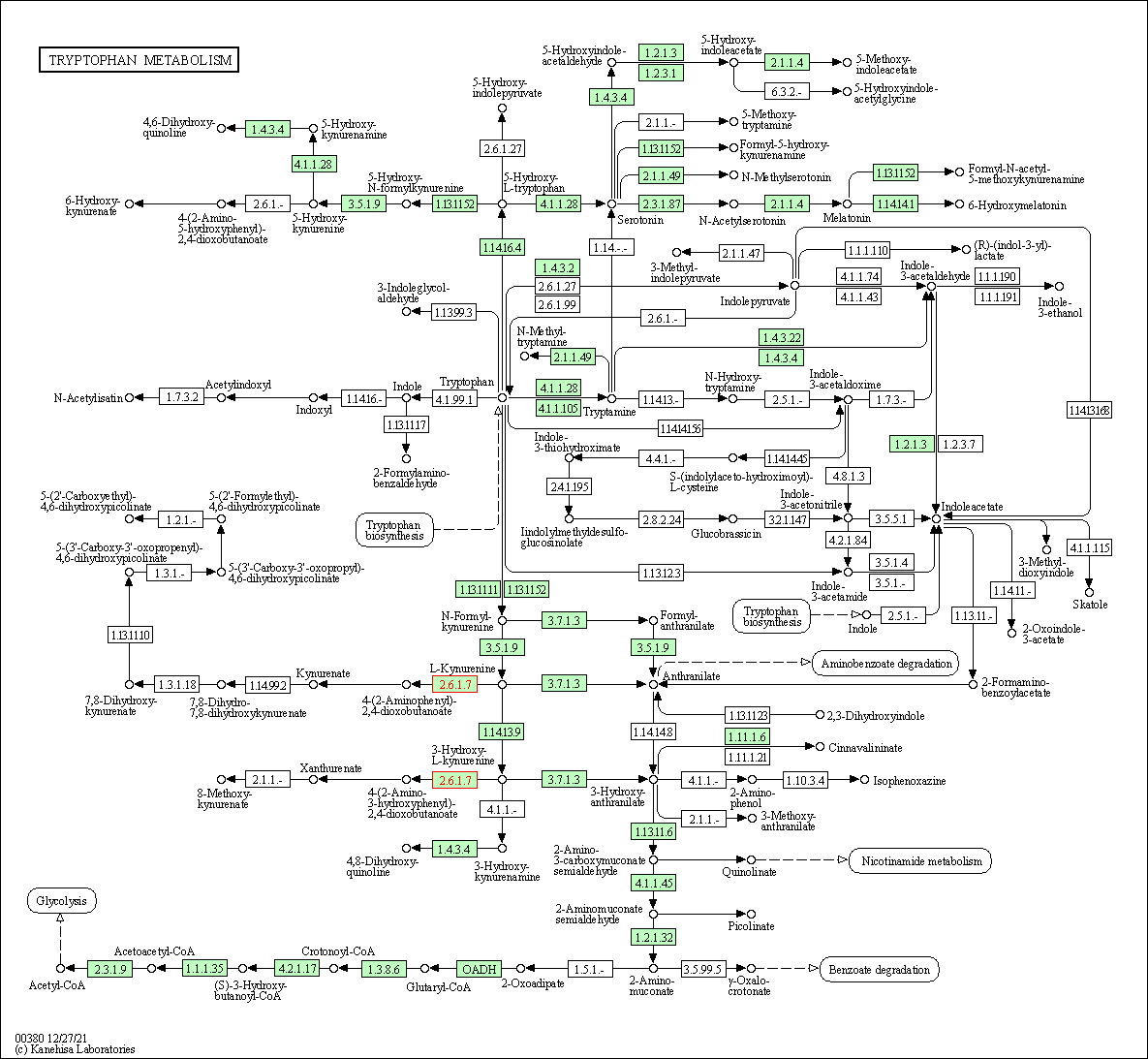
|
| Class: Metabolism => Amino acid metabolism | Pathway Hierarchy | ||
| Degree | 4 | Degree centrality | 4.30E-04 | Betweenness centrality | 0.00E+00 |
|---|---|---|---|---|---|
| Closeness centrality | 1.36E-01 | Radiality | 1.14E+01 | Clustering coefficient | 1.00E+00 |
| Neighborhood connectivity | 7.00E+00 | Topological coefficient | 6.36E-01 | Eccentricity | 14 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Improvement of kynurenine aminotransferase-II inhibitors guided by mimicking sulfate esters. PLoS One. 2018 Apr 24;13(4):e0196404. | |||||
| REF 2 | Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nat Rev Drug Discov. 2019 May;18(5):379-401. | |||||
| REF 3 | Bicyclic or tricyclic heterocyclic compound. US10065972. | |||||
| REF 4 | KAT II inhibitors. US8933095. | |||||
| REF 5 | KAT II inhibitors. US8598200. | |||||
| REF 6 | Specific inhibition of kynurenate synthesis enhances extracellular dopamine levels in the rodent striatum. Neuroscience. 2009 Mar 3;159(1):196-203. | |||||
| REF 7 | Investigating KYNA production and kynurenergic manipulation on acute mouse brain slice preparations. Brain Res Bull. 2019 Mar;146:185-191. | |||||
| REF 8 | Crystal structure-based selective targeting of the pyridoxal 5'-phosphate dependent enzyme kynurenine aminotransferase II for cognitive enhancement. J Med Chem. 2010 Aug 12;53(15):5684-9. | |||||
| REF 9 | Crystal structure of human kynurenine aminotransferase II. J Biol Chem. 2008 Feb 8;283(6):3567-3573. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

