Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T79307
(Former ID: TTDI03612)
|
|||||
| Target Name |
Serine/threonine-protein kinase ULK1 (ULK1)
|
|||||
| Synonyms |
hATG1; Unc-51-like kinase 1; KIAA0722; Autophagy-related protein 1 homolog; ATG1
Click to Show/Hide
|
|||||
| Gene Name |
ULK1
|
|||||
| Target Type |
Literature-reported target
|
[1] | ||||
| Function |
Acts upstream of phosphatidylinositol 3-kinase PIK3C3 to regulate the formation of autophagophores, the precursors of autophagosomes. Part of regulatory feedback loops in autophagy: acts both as a downstream effector and negative regulator of mammalian target of rapamycin complex 1 (mTORC1) via interaction with RPTOR. Activated via phosphorylation by AMPK and also acts as a regulator of AMPK by mediating phosphorylation of AMPK subunits PRKAA1, PRKAB2 and PRKAG1, leading to negatively regulate AMPK activity. May phosphorylate ATG13/KIAA0652 and RPTOR; however such data need additional evidences. Plays a role early in neuronal differentiation and is required for granule cell axon formation. May also phosphorylate SESN2 and SQSTM1 to regulate autophagy. Serine/threonine-protein kinase involved in autophagy in response to starvation.
Click to Show/Hide
|
|||||
| BioChemical Class |
Kinase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.7.11.1
|
|||||
| Sequence |
MEPGRGGTETVGKFEFSRKDLIGHGAFAVVFKGRHREKHDLEVAVKCINKKNLAKSQTLL
GKEIKILKELKHENIVALYDFQEMANSVYLVMEYCNGGDLADYLHAMRTLSEDTIRLFLQ QIAGAMRLLHSKGIIHRDLKPQNILLSNPAGRRANPNSIRVKIADFGFARYLQSNMMAAT LCGSPMYMAPEVIMSQHYDGKADLWSIGTIVYQCLTGKAPFQASSPQDLRLFYEKNKTLV PTIPRETSAPLRQLLLALLQRNHKDRMDFDEFFHHPFLDASPSVRKSPPVPVPSYPSSGS GSSSSSSSTSHLASPPSLGEMQQLQKTLASPADTAGFLHSSRDSGGSKDSSCDTDDFVMV PAQFPGDLVAEAPSAKPPPDSLMCSGSSLVASAGLESHGRTPSPSPPCSSSPSPSGRAGP FSSSRCGASVPIPVPTQVQNYQRIERNLQSPTQFQTPRSSAIRRSGSTSPLGFARASPSP PAHAEHGGVLARKMSLGGGRPYTPSPQVGTIPERPGWSGTPSPQGAEMRGGRSPRPGSSA PEHSPRTSGLGCRLHSAPNLSDLHVVRPKLPKPPTDPLGAVFSPPQASPPQPSHGLQSCR NLRGSPKLPDFLQRNPLPPILGSPTKAVPSFDFPKTPSSQNLLALLARQGVVMTPPRNRT LPDLSEVGPFHGQPLGPGLRPGEDPKGPFGRSFSTSRLTDLLLKAAFGTQAPDPGSTESL QEKPMEIAPSAGFGGSLHPGARAGGTSSPSPVVFTVGSPPSGSTPPQGPRTRMFSAGPTG SASSSARHLVPGPCSEAPAPELPAPGHGCSFADPITANLEGAVTFEAPDLPEETLMEQEH TEILRGLRFTLLFVQHVLEIAALKGSASEAAGGPEYQLQESVVADQISLLSREWGFAEQL VLYLKVAELLSSGLQSAIDQIRAGKLCLSSTVKQVVRRLNELYKASVVSCQGLSLRLQRF FLDKQRLLDRIHSITAERLIFSHAVQMVQSAALDEMFQHREGCVPRYHKALLLLEGLQHM LSDQADIENVTKCKLCIERRLSALLTGICA Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Phosphonothreonine | Ligand Info | |||||
| Structure Description | Structure of ULK1 bound to an inhibitor | PDB:4WNO | ||||
| Method | X-ray diffraction | Resolution | 1.56 Å | Mutation | Yes | [2] |
| PDB Sequence |
TETVGKFEFS
17 RKDLIGHGAF27 AVVFKGRHRA37 AHDLEVAVKC47 INKKNLAKSQ57 TLLGKEIKIL 67 KELKHENIVA77 LYDFQEMANS87 VYLVMEYCNG97 GDLADYLHAM107 RTLSEDTIRL 117 FLQQIAGAMR127 LLHSKGIIHR137 DLKPQNILLS147 NPAGRRANPN157 SIRVKIADFG 167 FARYLQSNMM177 AALCGSPMYM188 APEVIMSQHY198 DGKADLWSIG208 TIVYQCLTGK 218 APFQASSPQD228 LRLFYEKNKT238 LVPTIPRETS248 APLRQLLLAL258 LQRNHKDRMD 268 FDEFFHHPFL278 DA
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Ligand Name: Cysteine Sulfenic Acid | Ligand Info | |||||
| Structure Description | ULK1 Unc-51 like autophagy activating kinase in complex with inhibitor BTC | PDB:6MNH | ||||
| Method | X-ray diffraction | Resolution | 1.73 Å | Mutation | Yes | [3] |
| PDB Sequence |
TETVGKFEFS
17 RKDLIGHGAF27 AVVFKGRHRE37 KHDLEVAVKC47 INKKNLAKSQ57 TLLGKEIKIL 67 KELKHENIVA77 LYDFQEMANV88 YLVMEYCNGG98 DLADYLHAMR108 TLSEDTIRLF 118 LQQIAGAMRL128 LHSKGIIHRD138 LKPQNILLSN148 SIRVKIADFG167 FARYLTLGSP 185 MYMAPEVIMA195 QHYDGKADLW205 SIGTIVYQCL215 TGKAPFQAAS225 PQDLRLFYEK 235 NKTLVPTIPR245 ETSAPLRQLL255 LALLQRNHKD265 RMDFDEFFHH275 PFLDASPSVR 285 KSPPV
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Mitophagy - animal | hsa04137 | Affiliated Target |
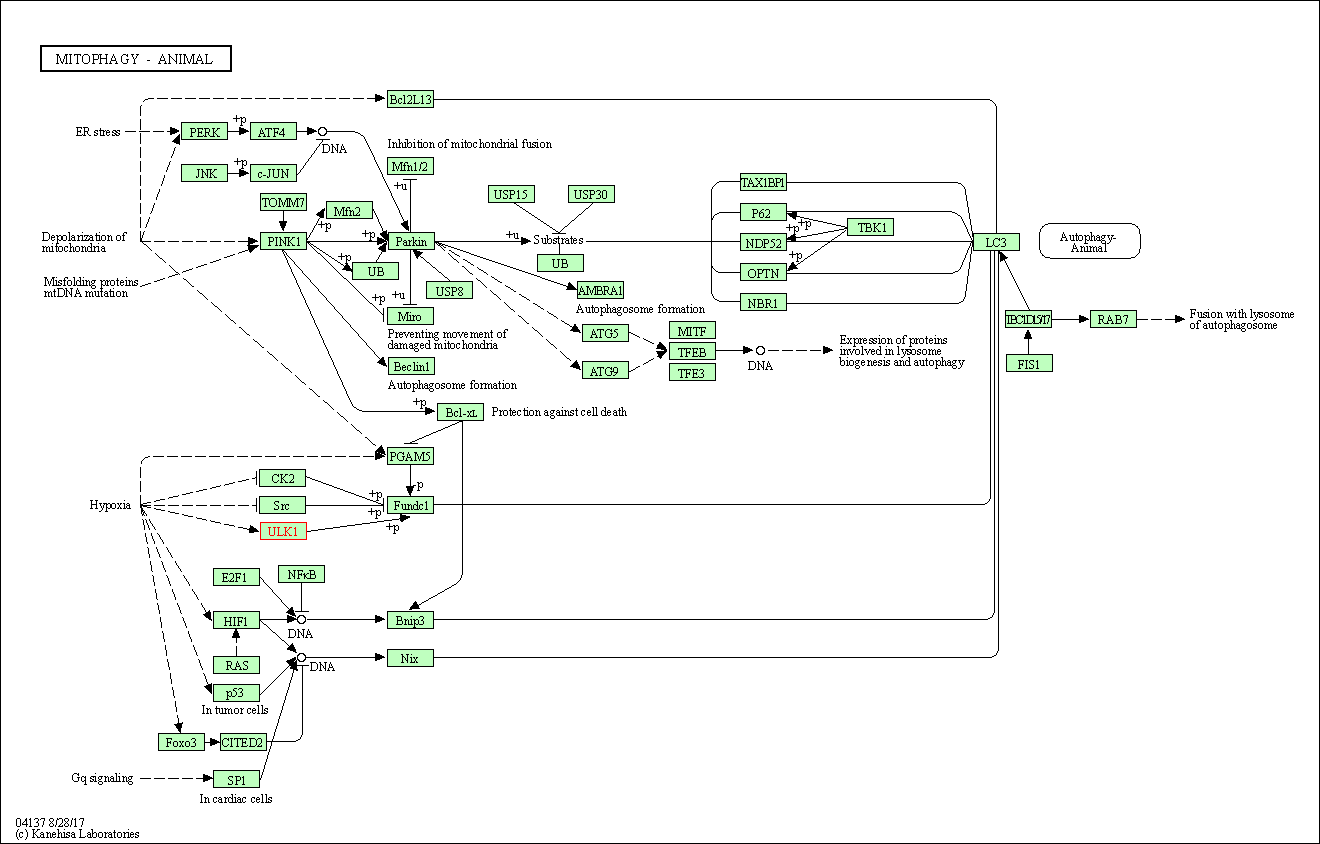
|
| Class: Cellular Processes => Transport and catabolism | Pathway Hierarchy | ||
| Autophagy - animal | hsa04140 | Affiliated Target |
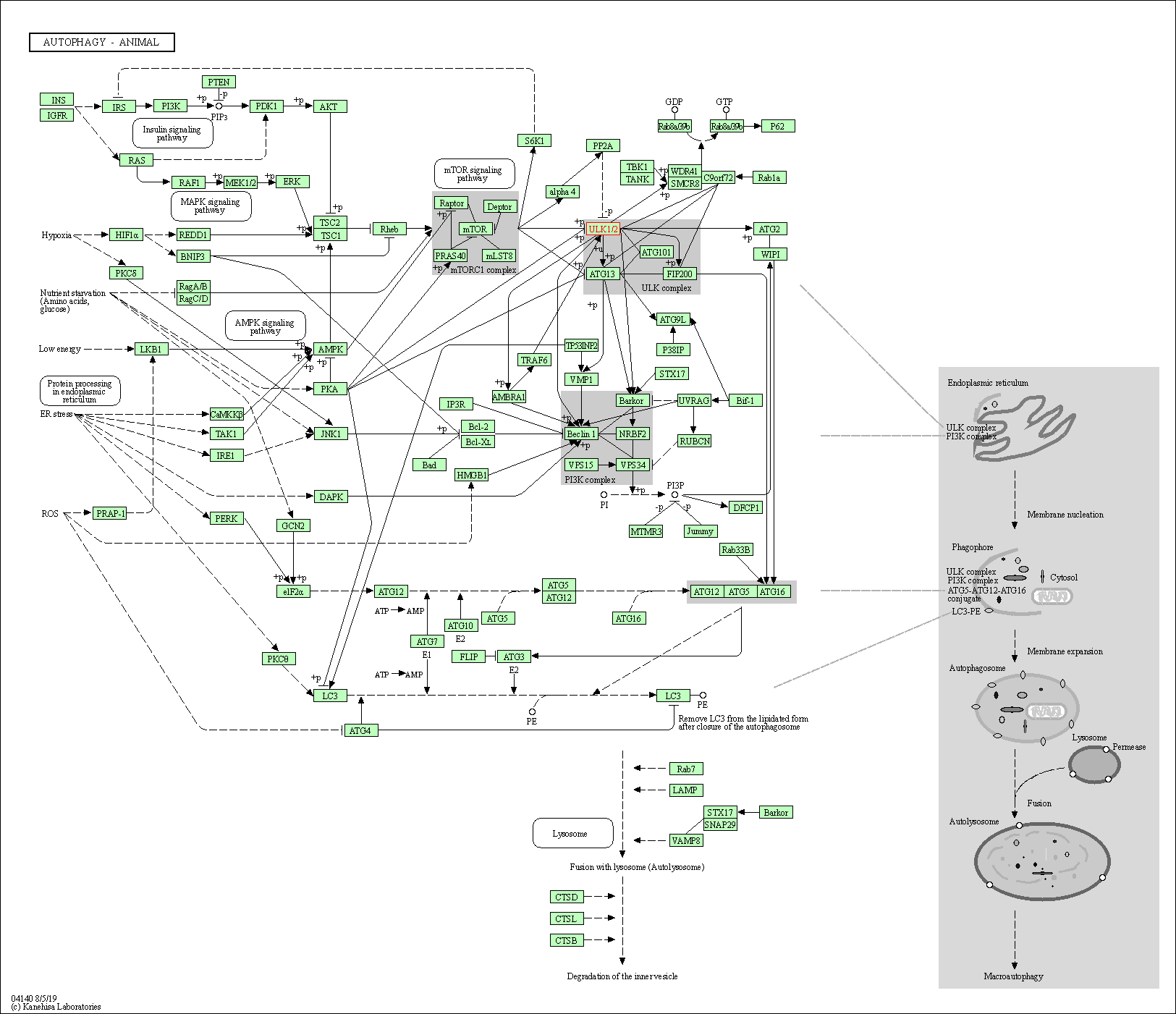
|
| Class: Cellular Processes => Transport and catabolism | Pathway Hierarchy | ||
| mTOR signaling pathway | hsa04150 | Affiliated Target |
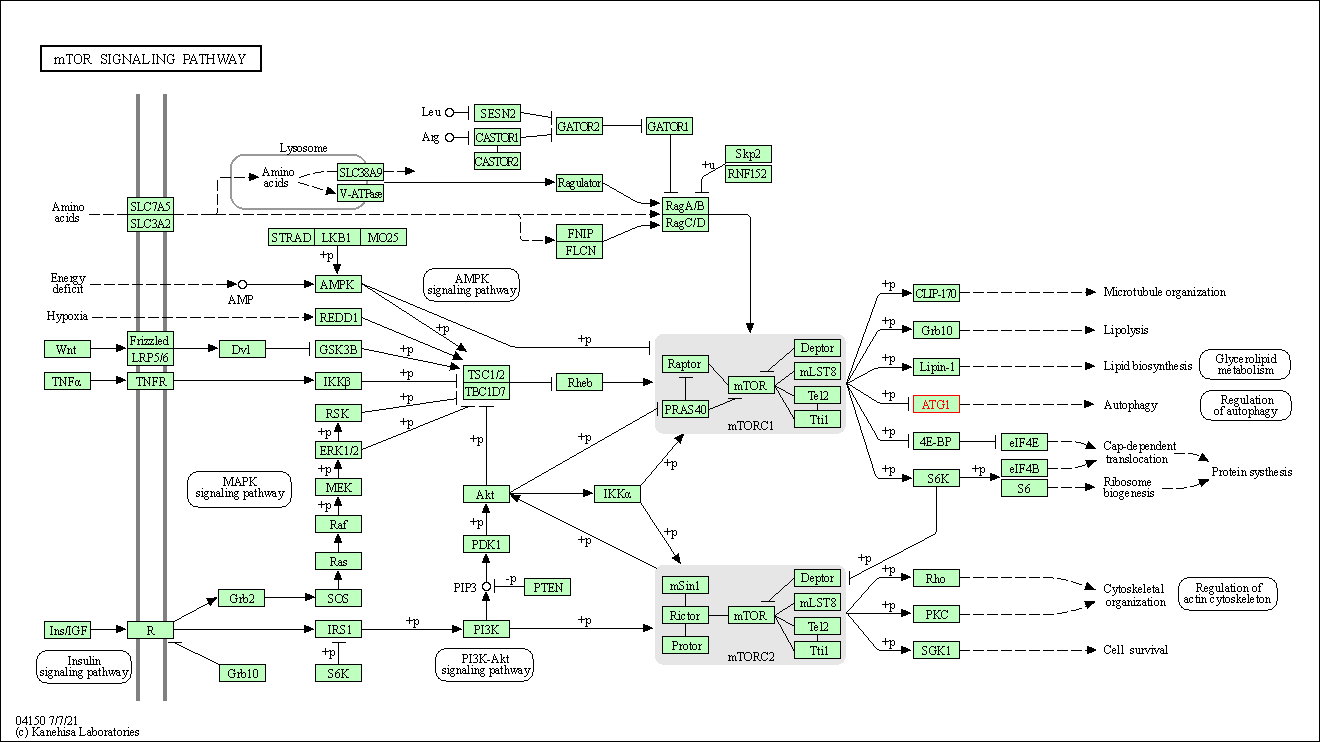
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| AMPK signaling pathway | hsa04152 | Affiliated Target |
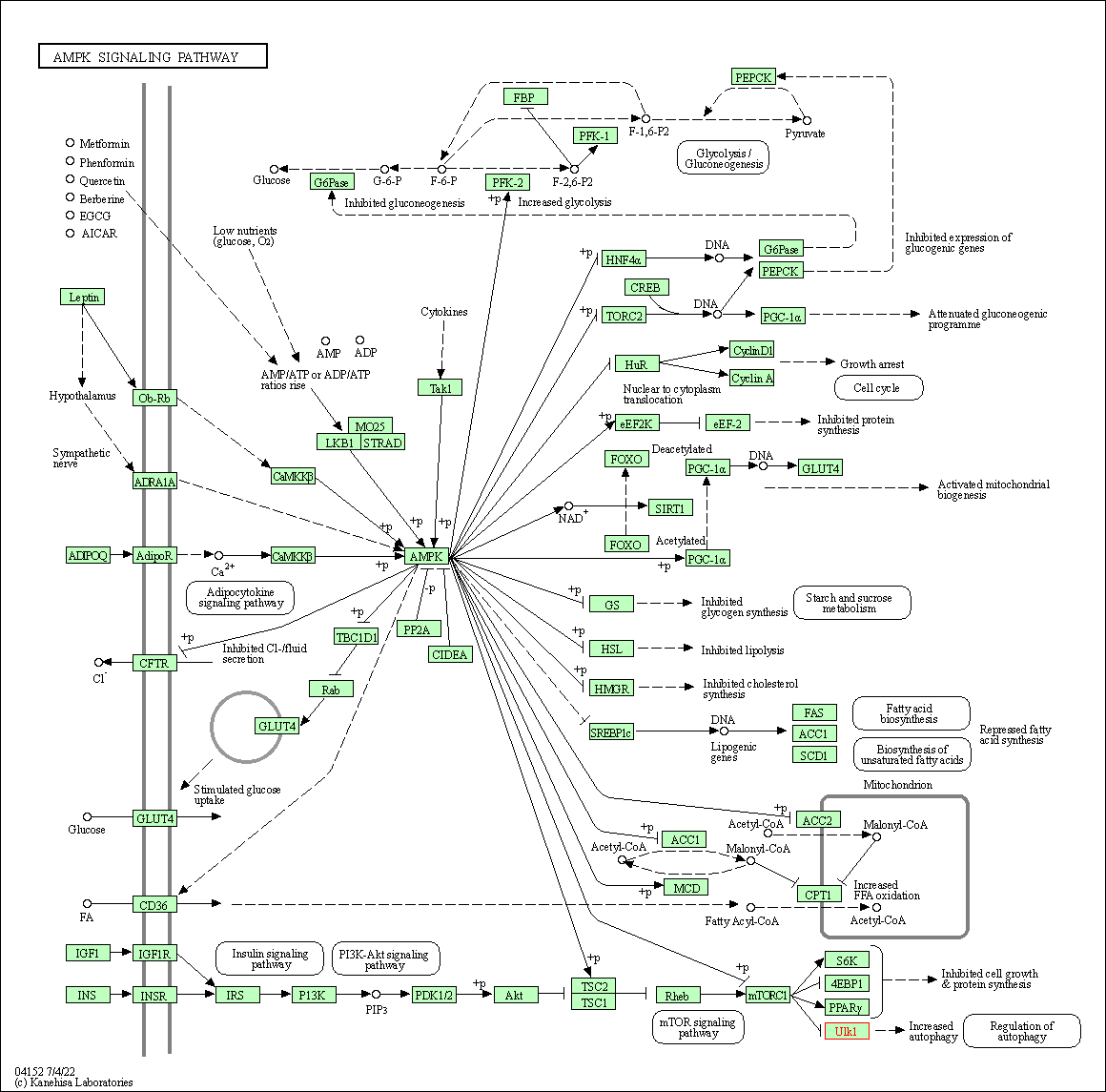
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Longevity regulating pathway | hsa04211 | Affiliated Target |
|
| Class: Organismal Systems => Aging | Pathway Hierarchy | ||
| Degree | 22 | Degree centrality | 2.36E-03 | Betweenness centrality | 9.70E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 2.25E-01 | Radiality | 1.40E+01 | Clustering coefficient | 4.20E-01 |
| Neighborhood connectivity | 2.68E+01 | Topological coefficient | 1.23E-01 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Improvement in oral bioavailability of 2,4-diaminopyrimidine c-Met inhibitors by incorporation of a 3-amidobenzazepin-2-one group. Bioorg Med Chem. 2011 Nov 1;19(21):6274-84. | |||||
| REF 2 | Structure of the human autophagy initiating kinase ULK1 in complex with potent inhibitors. ACS Chem Biol. 2015 Jan 16;10(1):257-61. | |||||
| REF 3 | Idea2Data: Toward a New Paradigm for Drug Discovery. ACS Med Chem Lett. 2019 Feb 4;10(3):278-286. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

