Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T78510
(Former ID: TTDI03122)
|
|||||
| Target Name |
Cyclic nucleotide-gated channel alpha-3 (CNGA3)
|
|||||
| Synonyms |
Cyclic nucleotide-gated cation channel alpha-3; Cone photoreceptor cGMP-gated channel subunit alpha; CNG3; CNG-3; CNG channel alpha-3; CNCG3
Click to Show/Hide
|
|||||
| Gene Name |
CNGA3
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Inherited retinal dystrophy [ICD-11: 9B70] | |||||
| Function |
Visual signal transduction is mediated by a G-protein coupled cascade using cGMP as second messenger. This protein can be activated by cyclic GMP which leads to an opening of the cation channel and thereby causing a depolarization of cone photoreceptors. Induced a flickering channel gating, weakened the outward rectification in the presence of extracellular calcium, increased sensitivity for L-cis diltiazem and enhanced the cAMP efficacy of the channel when coexpressed with CNGB3 (By similarity). Essential for the generation of light-evoked electrical responses in the red-, green- and blue sensitive cones.
Click to Show/Hide
|
|||||
| UniProt ID | ||||||
| Sequence |
MAKINTQYSHPSRTHLKVKTSDRDLNRAENGLSRAHSSSEETSSVLQPGIAMETRGLADS
GQGSFTGQGIARLSRLIFLLRRWAARHVHHQDQGPDSFPDRFRGAELKEVSSQESNAQAN VGSQEPADRGRSAWPLAKCNTNTSNNTEEEKKTKKKDAIVVDPSSNLYYRWLTAIALPVF YNWYLLICRACFDELQSEYLMLWLVLDYSADVLYVLDVLVRARTGFLEQGLMVSDTNRLW QHYKTTTQFKLDVLSLVPTDLAYLKVGTNYPEVRFNRLLKFSRLFEFFDRTETRTNYPNM FRIGNLVLYILIIIHWNACIYFAISKFIGFGTDSWVYPNISIPEHGRLSRKYIYSLYWST LTLTTIGETPPPVKDEEYLFVVVDFLVGVLIFATIVGNVGSMISNMNASRAEFQAKIDSI KQYMQFRKVTKDLETRVIRWFDYLWANKKTVDEKEVLKSLPDKLKAEIAINVHLDTLKKV RIFQDCEAGLLVELVLKLRPTVFSPGDYICKKGDIGKEMYIINEGKLAVVADDGVTQFVV LSDGSYFGEISILNIKGSKSGNRRTANIRSIGYSDLFCLSKDDLMEALTEYPEAKKALEE KGRQILMKDNLIDEELARAGADPKDLEEKVEQLGSSLDTLQTRFARLLAEYNATQMKMKQ RLSQLESQVKGGGDKPLADGEVPGDATKTEDKQQ Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| HIT2.0 ID | T68EUG | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 2 Clinical Trial Drugs | + | ||||
| 1 | AAV-CNGA3 | Drug Info | Phase 1/2 | Achromatopsia | [2] | |
| 2 | ACHM-CNGA3 | Drug Info | Phase 1/2 | Achromatopsia | [3] | |
| Mode of Action | [+] 1 Modes of Action | + | ||||
| Inhibitor | [+] 1 Inhibitor drugs | + | ||||
| 1 | L-(cis)-diltiazem | Drug Info | [1] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|

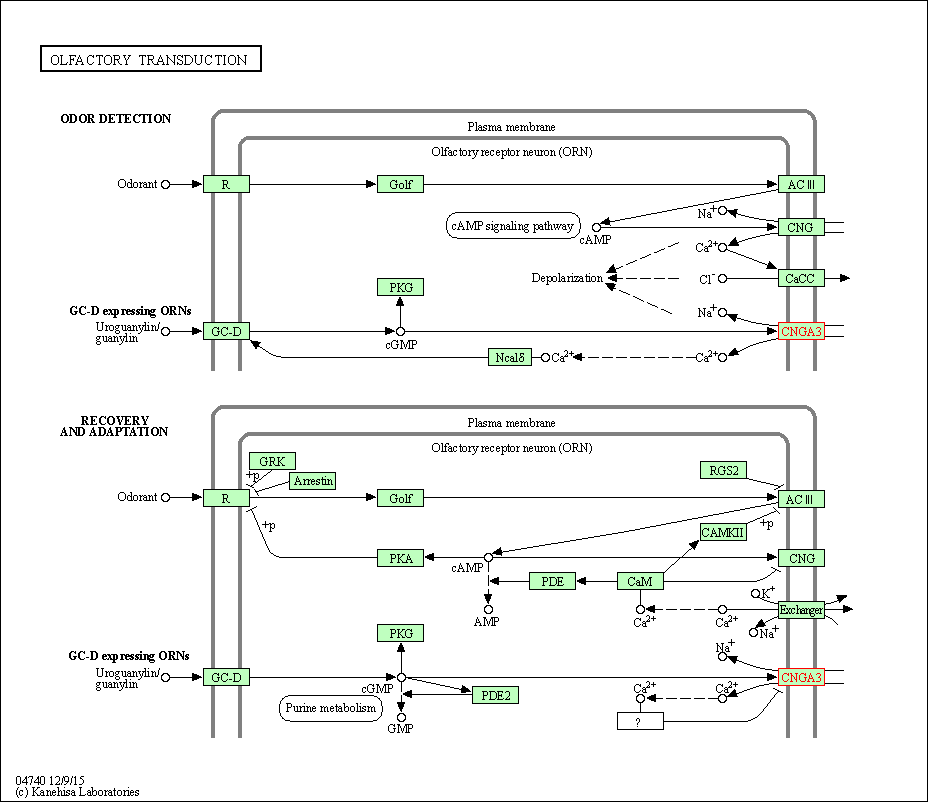
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| cAMP signaling pathway | hsa04024 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Olfactory transduction | hsa04740 | Affiliated Target |

|
| Class: Organismal Systems => Sensory system | Pathway Hierarchy | ||
| Degree | 4 | Degree centrality | 4.30E-04 | Betweenness centrality | 0.00E+00 |
|---|---|---|---|---|---|
| Closeness centrality | 1.60E-01 | Radiality | 1.24E+01 | Clustering coefficient | 1.00E+00 |
| Neighborhood connectivity | 6.50E+00 | Topological coefficient | 5.42E-01 | Eccentricity | 14 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Target id: 396). | |||||
| REF 2 | ClinicalTrials.gov (NCT03758404) Gene Therapy for Achromatopsia (CNGA3) (CNGA3). U.S. National Institutes of Health. | |||||
| REF 3 | Clinical pipeline report, company report or official report of Applied Genetic Technologies. | |||||
| REF 4 | Clinical pipeline report, company report or official report of MeiraGTx. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

