Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T47164
(Former ID: TTDI02447)
|
|||||
| Target Name |
MST-1 protein kinase (STK4)
|
|||||
| Synonyms |
Serine/threonine-protein kinase Krs-2; Serine/threonine-protein kinase 4 18kDa subunit; Serine/threonine-protein kinase 4; STE20-like kinase MST1; Mammalian STE20-like protein kinase 1; MST1/N; MST1/C; MST1; MST-1; KRS2
Click to Show/Hide
|
|||||
| Gene Name |
STK4
|
|||||
| Target Type |
Literature-reported target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Ischemic heart disease [ICD-11: BA40-BA6Z] | |||||
| Function |
Key component of the Hippo signaling pathway which plays a pivotal role in organ size control and tumor suppression by restricting proliferation and promoting apoptosis. The core of this pathway is composed of a kinase cascade wherein STK3/MST2 and STK4/MST1, in complex with its regulatory protein SAV1, phosphorylates and activates LATS1/2 in complex with its regulatory protein MOB1, which in turn phosphorylates and inactivates YAP1 oncoprotein and WWTR1/TAZ. Phosphorylation of YAP1 by LATS2 inhibits its translocation into the nucleus to regulate cellular genes important for cell proliferation, cell death, and cell migration. STK3/MST2 and STK4/MST1 are required to repress proliferation of mature hepatocytes, to prevent activation of facultative adult liver stem cells (oval cells), and to inhibit tumor formation. Phosphorylates 'Ser-14' of histone H2B (H2BS14ph) during apoptosis. Phosphorylates FOXO3 upon oxidative stress, which results in its nuclear translocation and cell death initiation. Phosphorylates MOBKL1A, MOBKL1B and RASSF2. Phosphorylates TNNI3 (cardiac Tn-I) and alters its binding affinity to TNNC1 (cardiac Tn-C) and TNNT2 (cardiac Tn-T). Phosphorylates FOXO1 on 'Ser-212' and regulates its activation and stimulates transcription of PMAIP1 in a FOXO1-dependent manner. Phosphorylates SIRT1 and inhibits SIRT1-mediated p53/TP53 deacetylation, thereby promoting p53/TP53 dependent transcription and apoptosis upon DNA damage. Acts as an inhibitor of PKB/AKT1. Phosphorylates AR on 'Ser-650' and suppresses its activity by intersecting with PKB/AKT1 signaling and antagonizing formation of AR-chromatin complexes. Stress-activated, pro-apoptotic kinase which, following caspase-cleavage, enters the nucleus and induces chromatin condensation followed by internucleosomal DNA fragmentation.
Click to Show/Hide
|
|||||
| BioChemical Class |
Kinase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.7.11.1
|
|||||
| Sequence |
METVQLRNPPRRQLKKLDEDSLTKQPEEVFDVLEKLGEGSYGSVYKAIHKETGQIVAIKQ
VPVESDLQEIIKEISIMQQCDSPHVVKYYGSYFKNTDLWIVMEYCGAGSVSDIIRLRNKT LTEDEIATILQSTLKGLEYLHFMRKIHRDIKAGNILLNTEGHAKLADFGVAGQLTDTMAK RNTVIGTPFWMAPEVIQEIGYNCVADIWSLGITAIEMAEGKPPYADIHPMRAIFMIPTNP PPTFRKPELWSDNFTDFVKQCLVKSPEQRATATQLLQHPFVRSAKGVSILRDLINEAMDV KLKRQESQQREVDQDDEENSEEDEMDSGTMVRAVGDEMGTVRVASTMTDGANTMIEHDDT LPSQLGTMVINAEDEEEEGTMKRRDETMQPAKPSFLEYFEQKEKENQINSFGKSVPGPLK NSSDWKIPQDGDYEFLKSWTVEDLQKRLLALDPMMEQEIEEIRQKYQSKRQPILDAIEAK KRRQQNF Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Phosphonothreonine | Ligand Info | |||||
| Structure Description | Crystal structure of Human STE20-like kinase 1, MST1 in complex with compound XMU-MP-1 | PDB:8A5J | ||||
| Method | X-ray diffraction | Resolution | 2.12 Å | Mutation | No | [2] |
| PDB Sequence |
EVFDVLEKLG
136 EGSVYKAIHK149 ETGQIVAIKQ159 VPVESDLQEI169 IKEISIMQQC179 DSPHVVKYYG 189 SYFKNTDLWI199 VMEYCGAGSV209 SDIIRLRNKT219 LTEDEIATIL229 QSTLKGLEYL 239 HFMRKIHRDI249 KAGNILLNTE259 GHAKLADFGV269 AGQLTDMAKR280 NVIGTPFWMA 291 PEVIQEIGYN301 CVADIWSLGI311 TAIEMAEGKP321 PYADIHPMRA331 IFMIPTNPPP 341 TFRKPELWSD351 NFTDFVKQCL361 VKSPEQRATA371 TQLLQHPFVR381 SAKGVSILRD 391 LINEAMDVKL401 KRQESQQ
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Ligand Name: (2R)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid | Ligand Info | |||||
| Structure Description | Crystal structure of STK4 (MST1) in complex with compound 6 | PDB:6YAT | ||||
| Method | X-ray diffraction | Resolution | 2.58 Å | Mutation | No | [3] |
| PDB Sequence |
DEDSLTKQPE
27 EVFDVLEKLG37 EGSYGSVYKA47 IHKETGQIVA57 IKQVPVESDL67 QEIIKEISIM 77 QQCDSPHVVK87 YYGSYFKNTD97 LWIVMEYCGA107 GSVSDIIRLR117 NKTLTEDEIA 127 TILQSTLKGL137 EYLHFMRKIH147 RDIKAGNILL157 NTEGHAKLAD167 FGVAGQLTDM 178 AKRNVIGTPF189 WMAPEVIQEI199 GYNCVADIWS209 LGITAIEMAE219 GKPPYADIHP 229 MRAIFMIPTN239 PPPTFRKPEL249 WSDNFTDFVK259 QCLVKSPEQR269 ATATQLLQHP 279 FVRSAKGVSI289 LRDLINEAMD299 VKLKRQESQ
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| MAPK signaling pathway | hsa04010 | Affiliated Target |
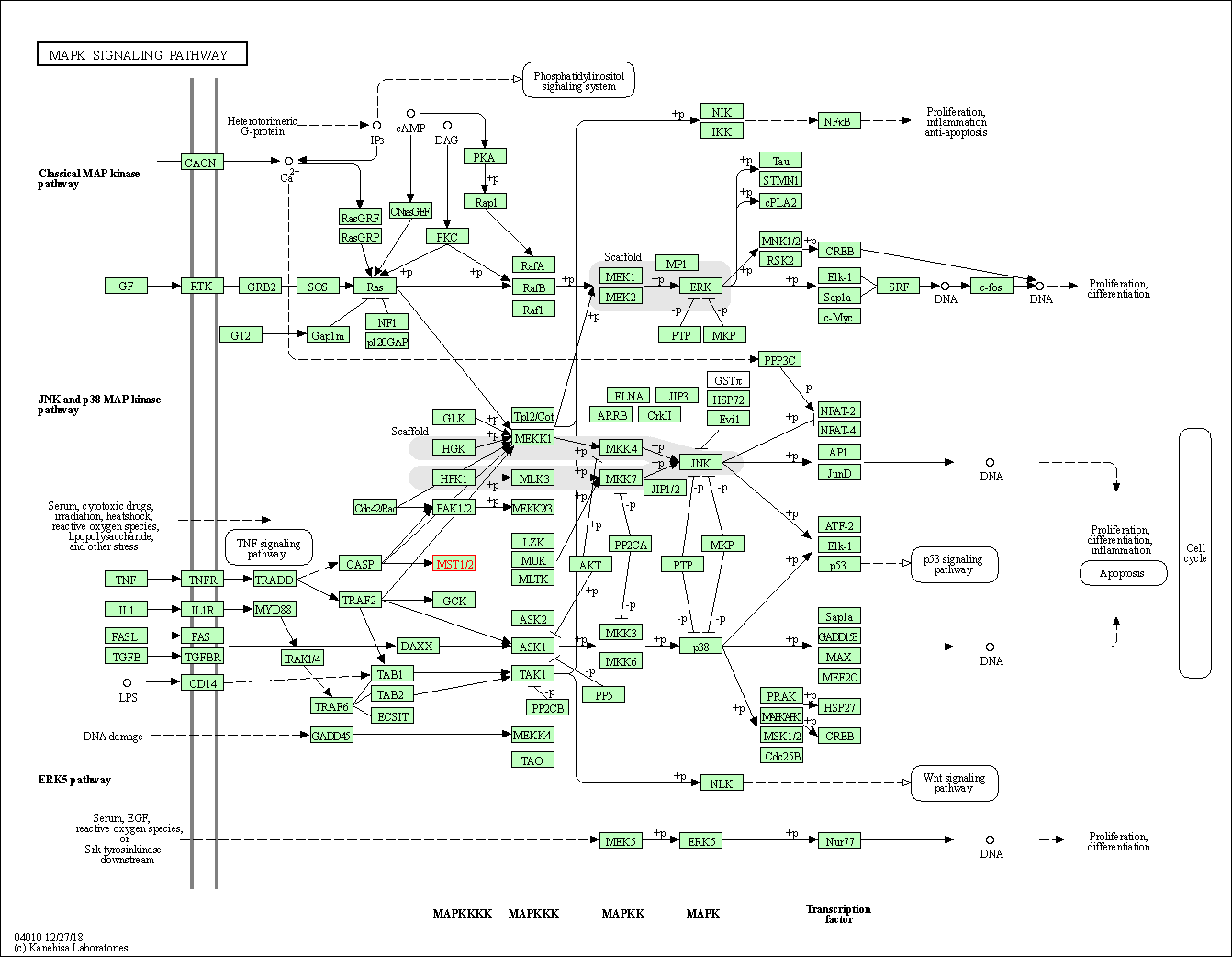
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Ras signaling pathway | hsa04014 | Affiliated Target |
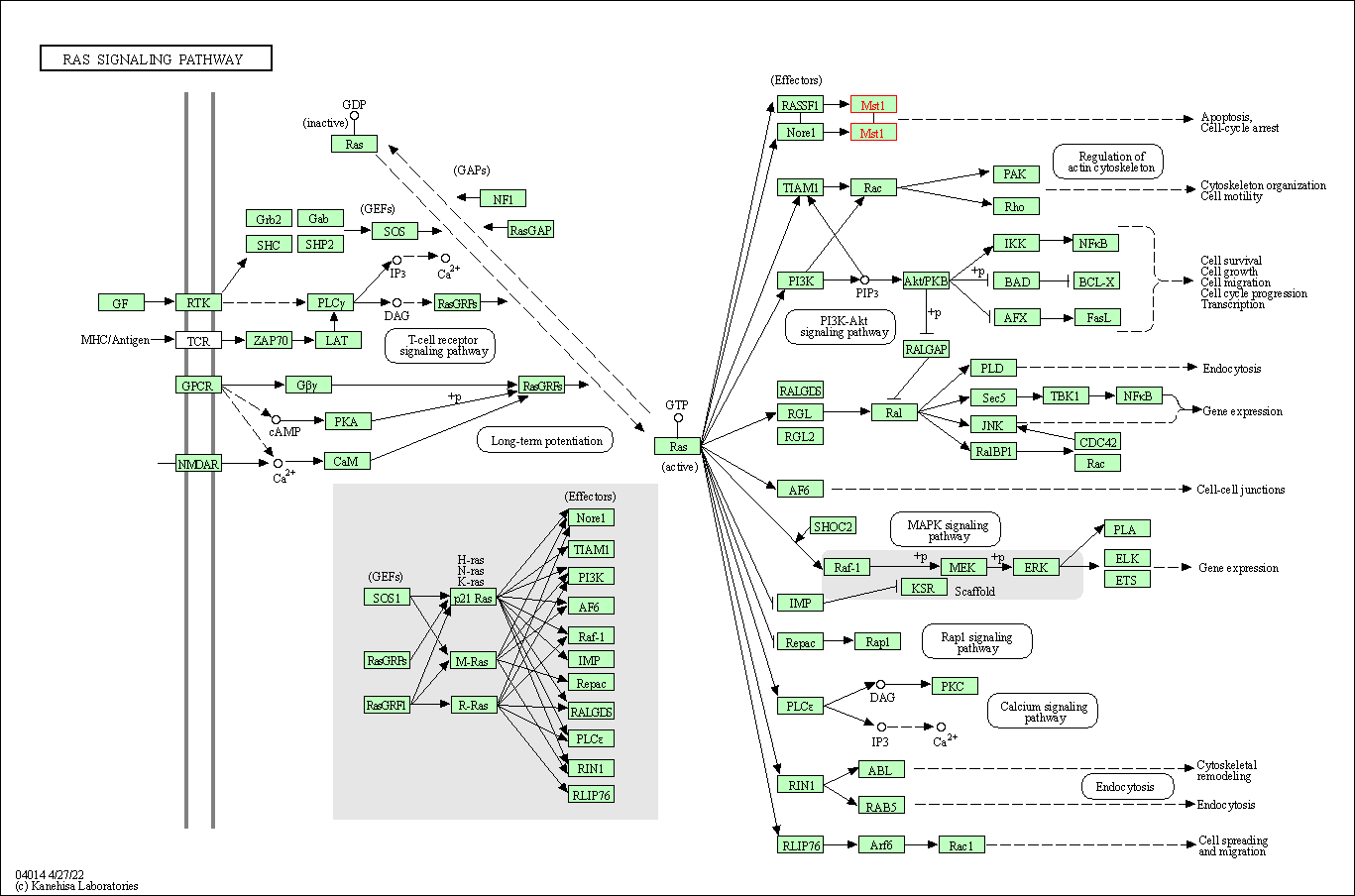
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| FoxO signaling pathway | hsa04068 | Affiliated Target |
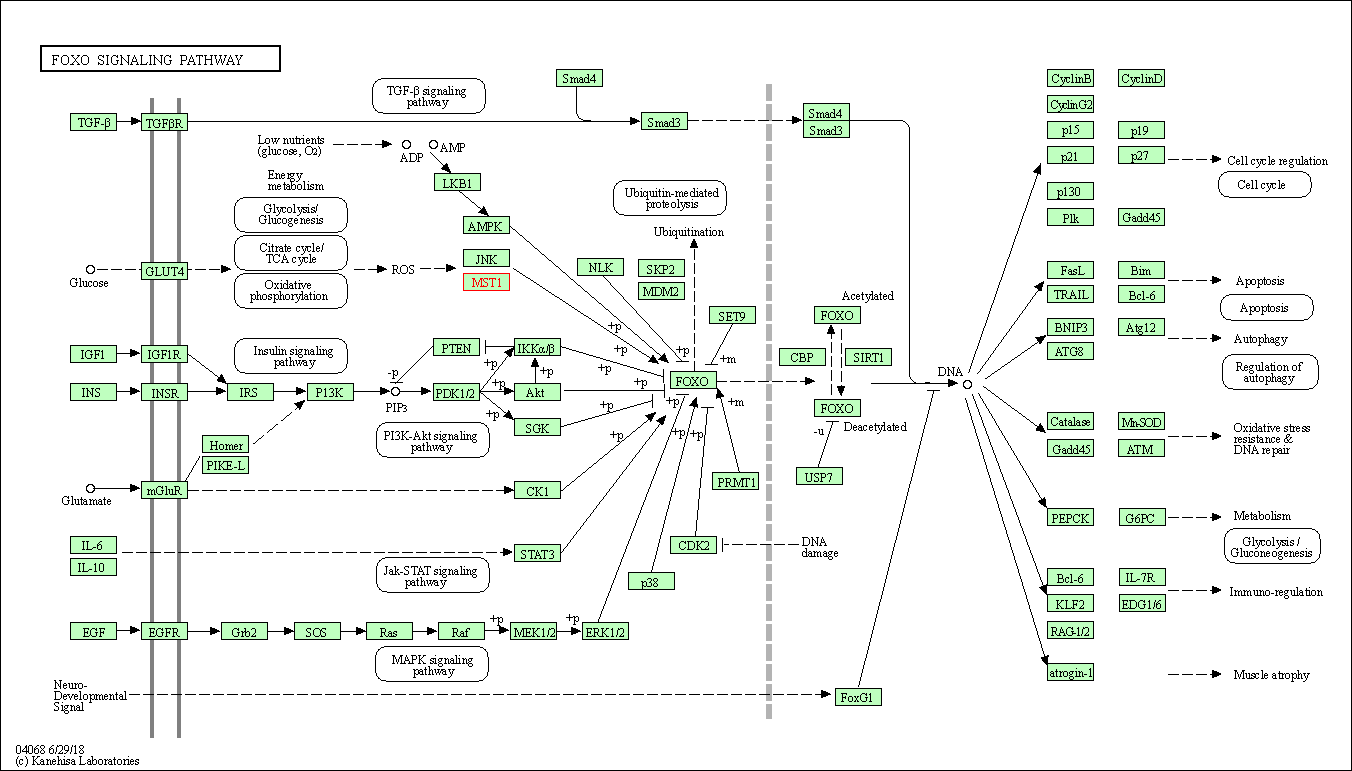
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Degree | 11 | Degree centrality | 1.18E-03 | Betweenness centrality | 1.88E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 2.09E-01 | Radiality | 1.37E+01 | Clustering coefficient | 4.36E-01 |
| Neighborhood connectivity | 1.52E+01 | Topological coefficient | 1.49E-01 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 5 KEGG Pathways | + | ||||
| 1 | MAPK signaling pathway | |||||
| 2 | Ras signaling pathway | |||||
| 3 | FoxO signaling pathway | |||||
| 4 | Pathways in cancer | |||||
| 5 | Non-small cell lung cancer | |||||
| NetPath Pathway | [+] 2 NetPath Pathways | + | ||||
| 1 | TSH Signaling Pathway | |||||
| 2 | TCR Signaling Pathway | |||||
| WikiPathways | [+] 1 WikiPathways | + | ||||
| 1 | Signaling by Hippo | |||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Target id: 2225). | |||||
| REF 2 | Crystal structure of the Kelch domain of human Keap1in complex with ligand S217879 | |||||
| REF 3 | Inhibitors of the Hippo Pathway Kinases STK3/MST2 and STK4/MST1 Have Utility for the Treatment of Acute Myeloid Leukemia. J Med Chem. 2022 Jan 27;65(2):1352-1369. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

