Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T35445
(Former ID: TTDR00819)
|
|||||
| Target Name |
Mothers against decapentaplegic homolog 3 (SMAD3)
|
|||||
| Synonyms |
TGF-beta response effector Smad3; Smad3; SMAD family member 3; SMAD 3; Mothers against DPP homolog 3; Mad3; MMad3; MADH3; MAD homolog 3; JV15-2; HSMAD3; HMAD-3
Click to Show/Hide
|
|||||
| Gene Name |
SMAD3
|
|||||
| Target Type |
Preclinical target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Kidney fibrosis [ICD-11: GC01] | |||||
| Function |
Binds the TRE element in the promoter region of many genes that are regulated by TGF-beta and, on formation of the SMAD3/SMAD4 complex, activates transcription. Also can form a SMAD3/SMAD4/JUN/FOS complex at the AP-1/SMAD site to regulate TGF-beta-mediated transcription. Has an inhibitory effect on wound healing probably by modulating both growth and migration of primary keratinocytes and by altering the TGF-mediated chemotaxis of monocytes. This effect on wound healing appears to be hormone-sensitive. Regulator of chondrogenesis and osteogenesis and inhibits early healing of bone fractures. Positively regulates PDPK1 kinase activity by stimulating its dissociation from the 14-3-3 protein YWHAQ which acts as a negative regulator. Receptor-regulated SMAD (R-SMAD) that is an intracellular signal transducer and transcriptional modulator activated by TGF-beta (transforming growth factor) and activin type 1 receptor kinases.
Click to Show/Hide
|
|||||
| UniProt ID | ||||||
| Sequence |
MSSILPFTPPIVKRLLGWKKGEQNGQEEKWCEKAVKSLVKKLKKTGQLDELEKAITTQNV
NTKCITIPRSLDGRLQVSHRKGLPHVIYCRLWRWPDLHSHHELRAMELCEFAFNMKKDEV CVNPYHYQRVETPVLPPVLVPRHTEIPAEFPPLDDYSHSIPENTNFPAGIEPQSNIPETP PPGYLSEDGETSDHQMNHSMDAGSPNLSPNPMSPAHNNLDLQPVTYCEPAFWCSISYYEL NQRVGETFHASQPSMTVDGFTDPSNSERFCLGLLSNVNRNAAVELTRRHIGRGVRLYYIG GEVFAECLSDSAIFVQSPNCNQRYGWHPATVCKIPPGCNLKIFNNQEFAALLAQSVNQGF EAVYQLTRMCTIRMSFVKGWGAEYRRQTVTSTPCWIELHLNGPLQWLDKVLTQMGSPSIR CSSVS Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| HIT2.0 ID | T78O6C | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Preclinical Drug(s) | [+] 1 Preclinical Drugs | + | ||||
| 1 | SIS-3 | Drug Info | Preclinical | Renal fibrosis | [2] | |
| Mode of Action | [+] 1 Modes of Action | + | ||||
| Inhibitor | [+] 3 Inhibitor drugs | + | ||||
| 1 | SIS-3 | Drug Info | [3] | |||
| 2 | ELLAGIC ACID | Drug Info | [1] | |||
| 3 | GNF-PF-2272 | Drug Info | [1] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| FoxO signaling pathway | hsa04068 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Cell cycle | hsa04110 | Affiliated Target |

|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Endocytosis | hsa04144 | Affiliated Target |

|
| Class: Cellular Processes => Transport and catabolism | Pathway Hierarchy | ||
| Cellular senescence | hsa04218 | Affiliated Target |

|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
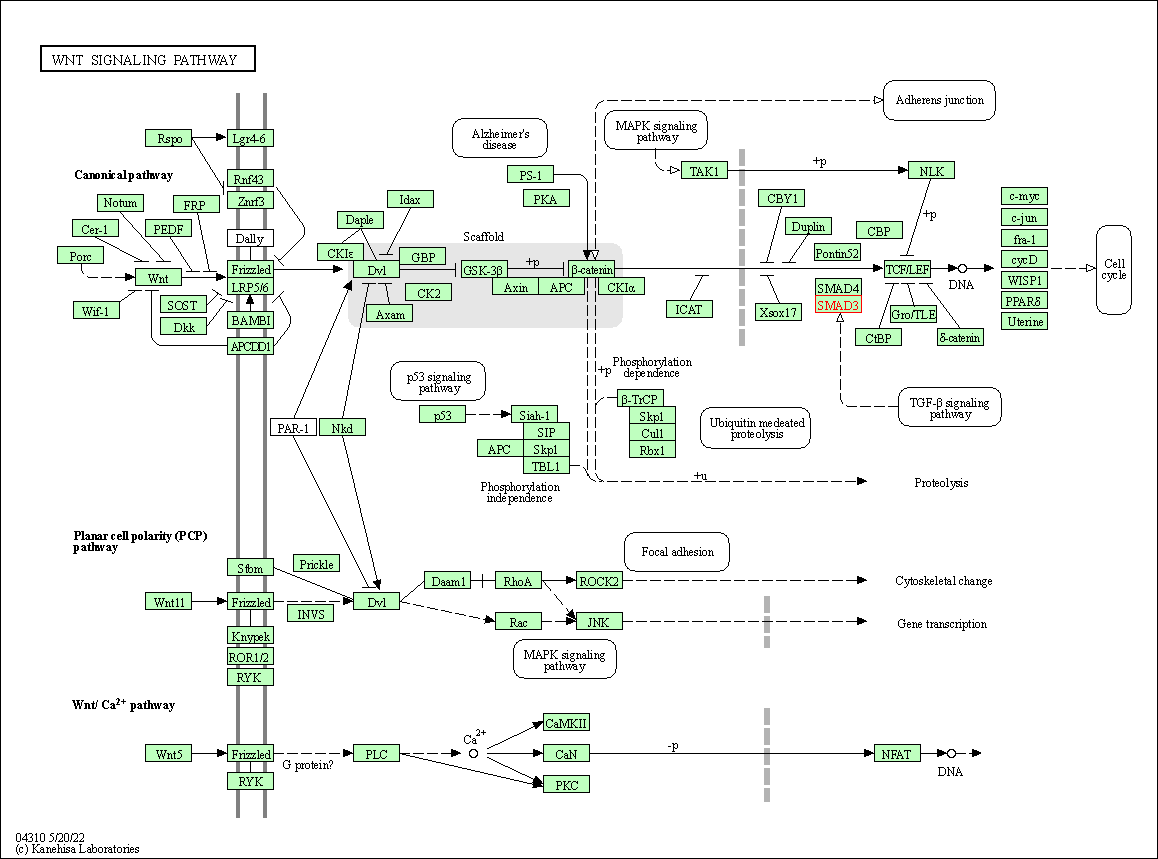
| Wnt signaling pathway | hsa04310 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
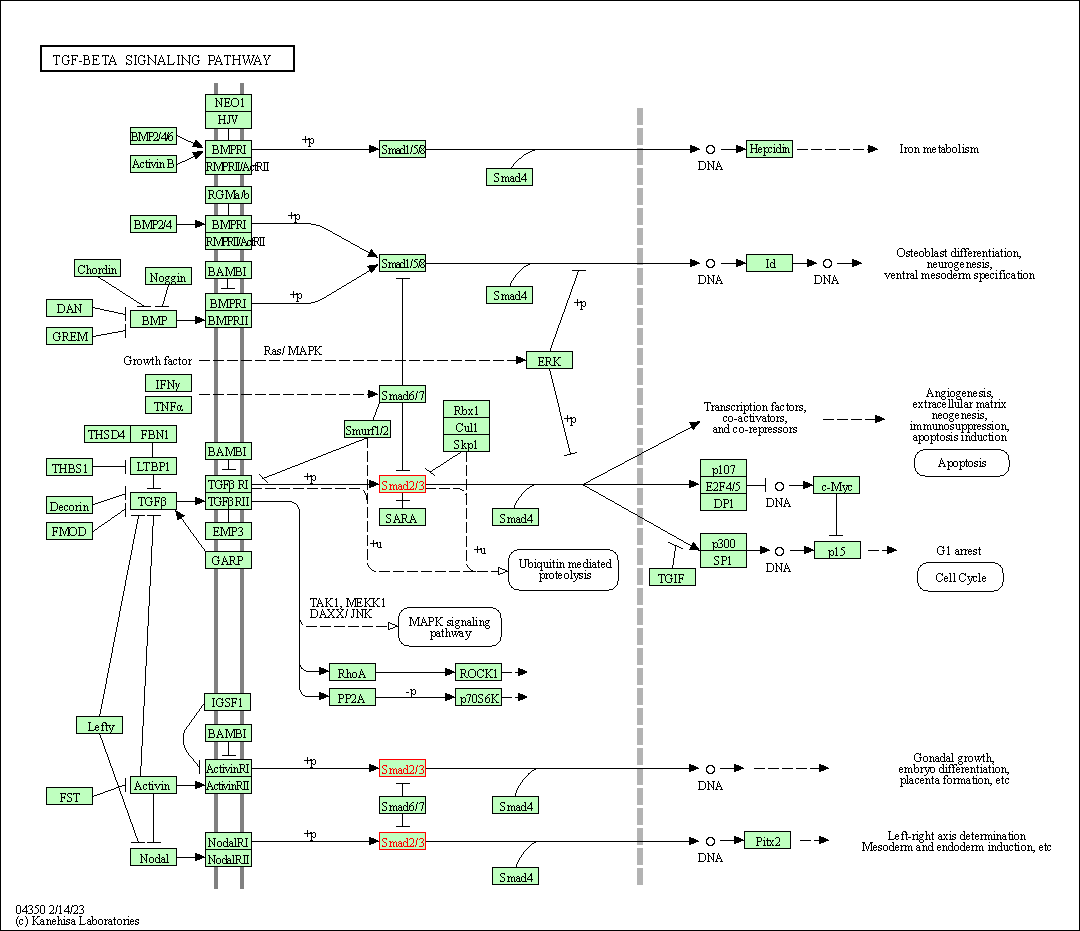
| TGF-beta signaling pathway | hsa04350 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
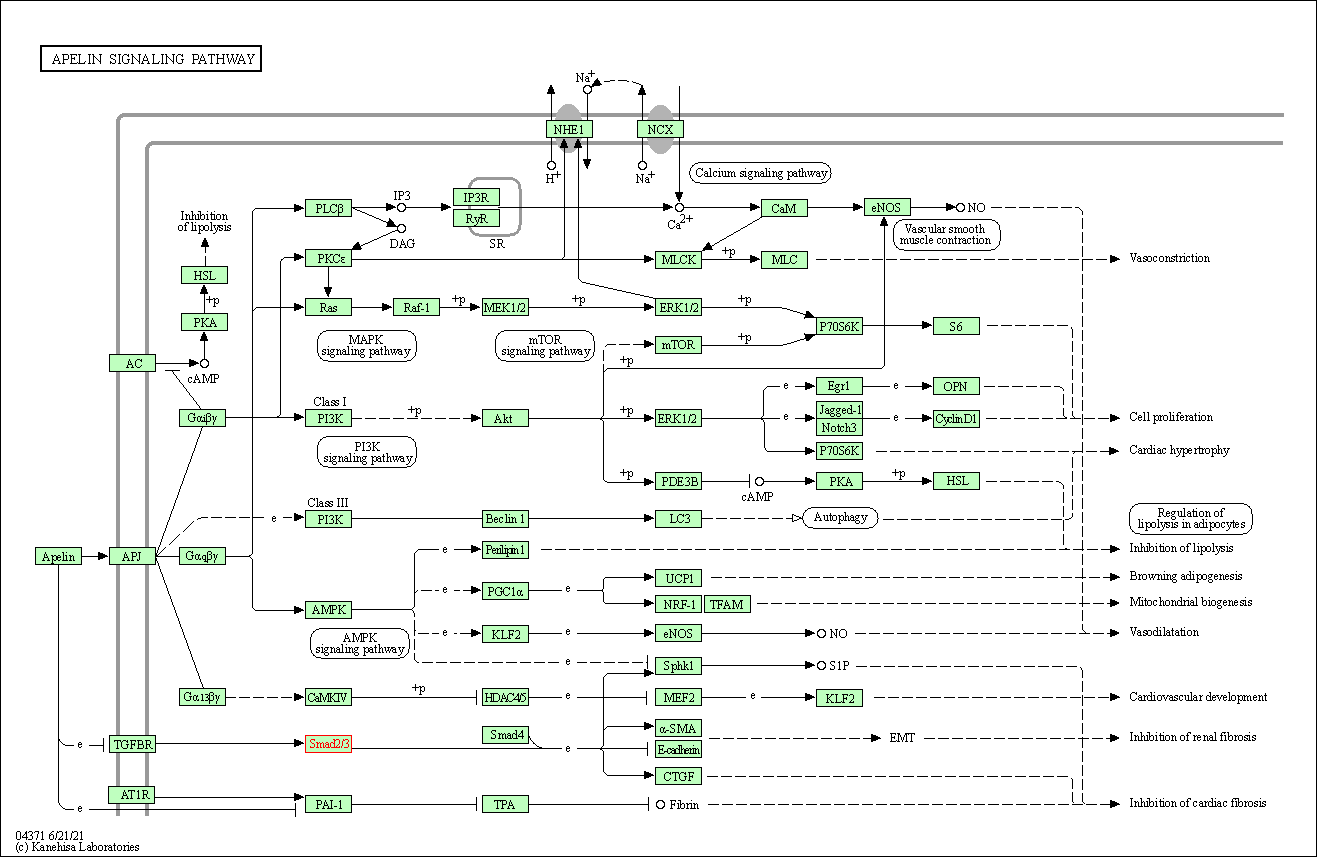
| Apelin signaling pathway | hsa04371 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
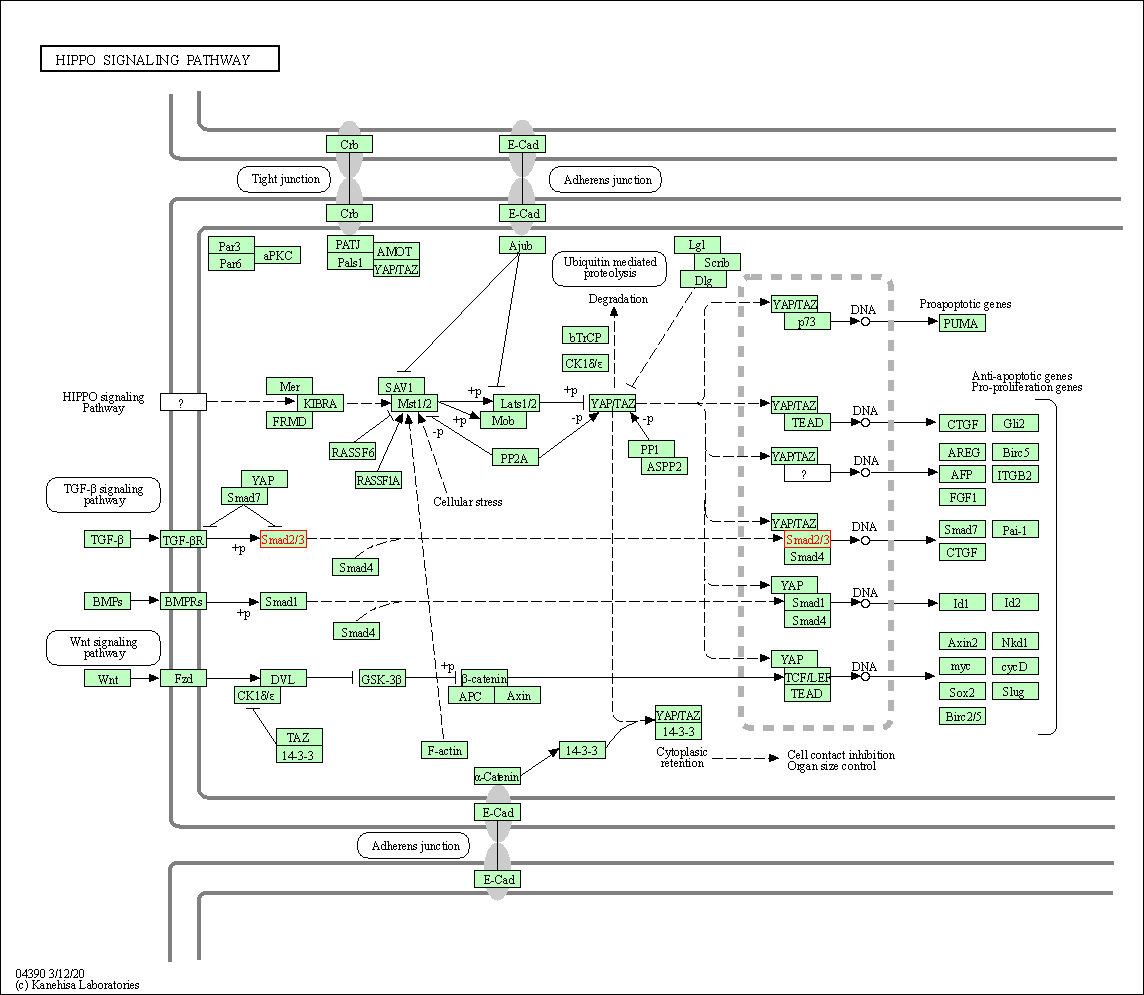
| Hippo signaling pathway | hsa04390 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
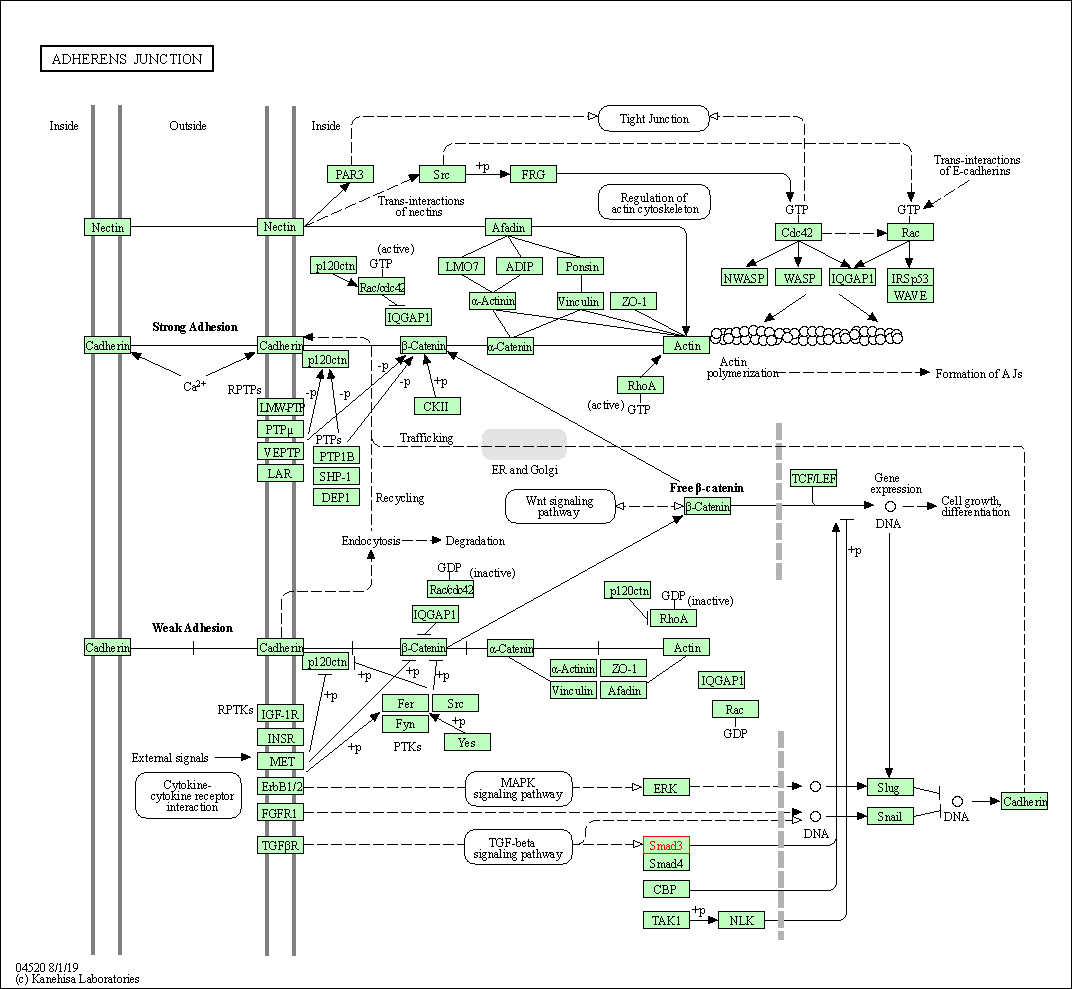
| Adherens junction | hsa04520 | Affiliated Target |

|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
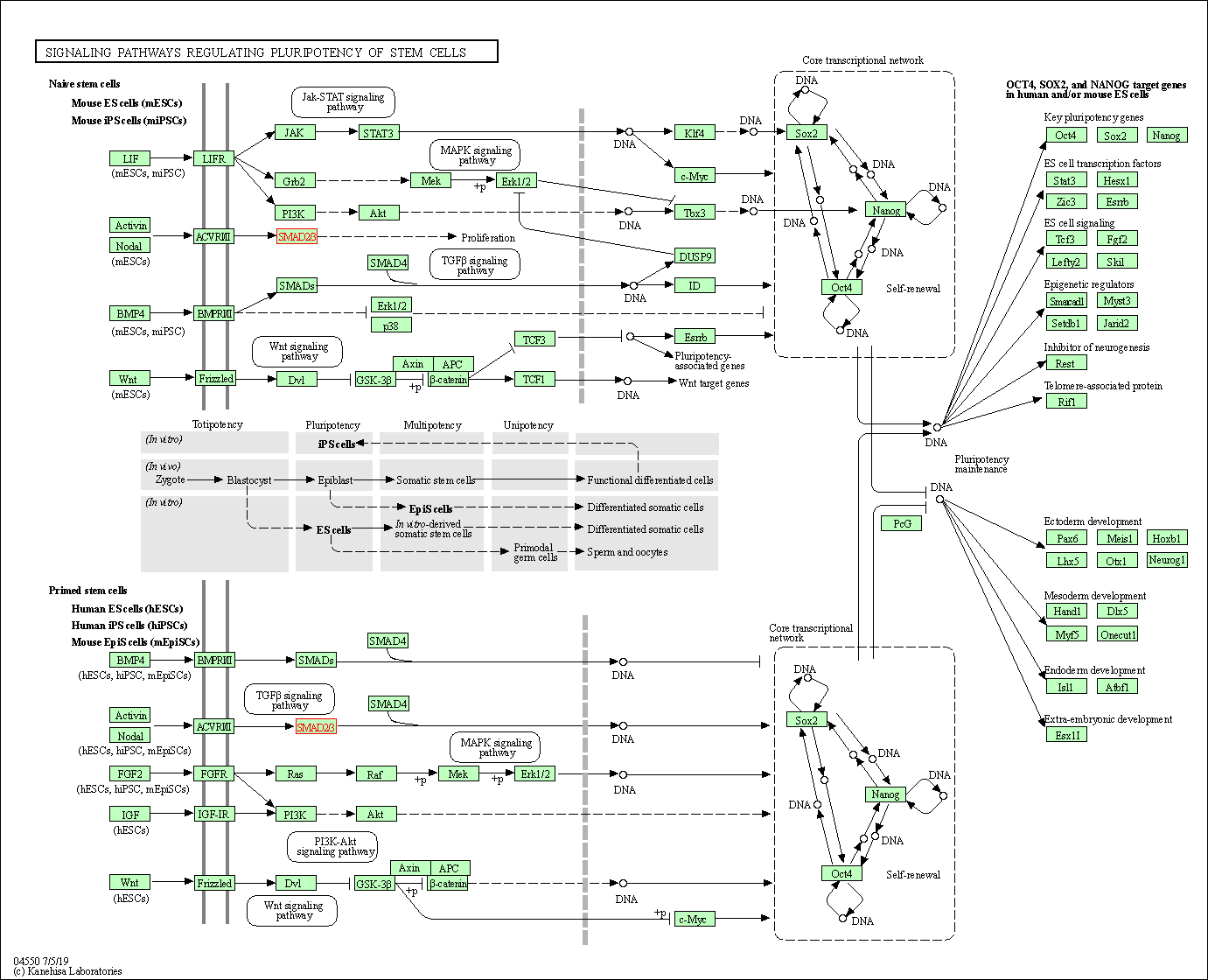
| Signaling pathways regulating pluripotency of stem cells | hsa04550 | Affiliated Target |

|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
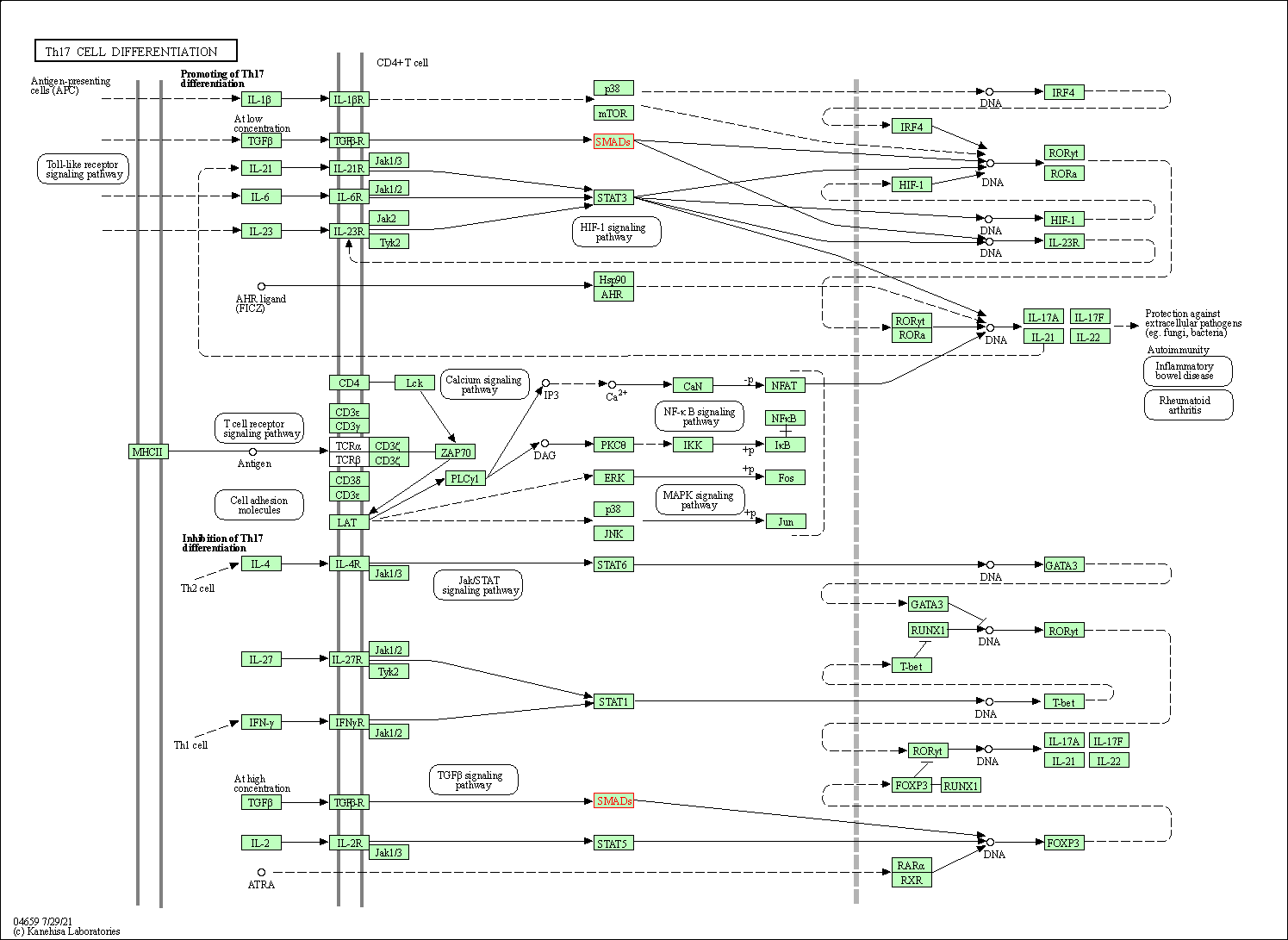
| Th17 cell differentiation | hsa04659 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 91 | Degree centrality | 9.78E-03 | Betweenness centrality | 7.50E-03 |
|---|---|---|---|---|---|
| Closeness centrality | 2.69E-01 | Radiality | 1.46E+01 | Clustering coefficient | 9.84E-02 |
| Neighborhood connectivity | 3.88E+01 | Topological coefficient | 2.80E-02 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | In silico identification and biochemical evaluation of novel inhibitors of NRH:quinone oxidoreductase 2 (NQO2). Bioorg Med Chem Lett. 2010 Dec 15;20(24):7331-6. | |||||
| REF 2 | Drugs and Targets in Fibrosis. Front Pharmacol. 2017 Nov 23;8:855. | |||||
| REF 3 | Characterization of SIS3, a novel specific inhibitor of Smad3, and its effect on transforming growth factor-beta1-induced extracellular matrix expression. Mol Pharmacol. 2006 Feb;69(2):597-607. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

