Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T33969
(Former ID: TTDS00278)
|
|||||
| Target Name |
ATP-binding cassette transporter B11 (ABCB11)
|
|||||
| Synonyms |
BSEP; ATP-binding cassette, sub-family B, member 11; ATP-binding cassette sub-family B member 11
Click to Show/Hide
|
|||||
| Gene Name |
ABCB11
|
|||||
| Target Type |
Literature-reported target
|
[1] | ||||
| Function |
Involved in the ATP-dependent secretion of bile salts into the canaliculus of hepatocytes.
Click to Show/Hide
|
|||||
| BioChemical Class |
ABC transporter
|
|||||
| UniProt ID | ||||||
| Sequence |
MSDSVILRSIKKFGEENDGFESDKSYNNDKKSRLQDEKKGDGVRVGFFQLFRFSSSTDIW
LMFVGSLCAFLHGIAQPGVLLIFGTMTDVFIDYDVELQELQIPGKACVNNTIVWTNSSLN QNMTNGTRCGLLNIESEMIKFASYYAGIAVAVLITGYIQICFWVIAAARQIQKMRKFYFR RIMRMEIGWFDCNSVGELNTRFSDDINKINDAIADQMALFIQRMTSTICGFLLGFFRGWK LTLVIISVSPLIGIGAATIGLSVSKFTDYELKAYAKAGVVADEVISSMRTVAAFGGEKRE VERYEKNLVFAQRWGIRKGIVMGFFTGFVWCLIFLCYALAFWYGSTLVLDEGEYTPGTLV QIFLSVIVGALNLGNASPCLEAFATGRAAATSIFETIDRKPIIDCMSEDGYKLDRIKGEI EFHNVTFHYPSRPEVKILNDLNMVIKPGEMTALVGPSGAGKSTALQLIQRFYDPCEGMVT VDGHDIRSLNIQWLRDQIGIVEQEPVLFSTTIAENIRYGREDATMEDIVQAAKEANAYNF IMDLPQQFDTLVGEGGGQMSGGQKQRVAIARALIRNPKILLLDMATSALDNESEAMVQEV LSKIQHGHTIISVAHRLSTVRAADTIIGFEHGTAVERGTHEELLERKGVYFTLVTLQSQG NQALNEEDIKDATEDDMLARTFSRGSYQDSLRASIRQRSKSQLSYLVHEPPLAVVDHKST YEEDRKDKDIPVQEEVEPAPVRRILKFSAPEWPYMLVGSVGAAVNGTVTPLYAFLFSQIL GTFSIPDKEEQRSQINGVCLLFVAMGCVSLFTQFLQGYAFAKSGELLTKRLRKFGFRAML GQDIAWFDDLRNSPGALTTRLATDASQVQGAAGSQIGMIVNSFTNVTVAMIIAFSFSWKL SLVILCFFPFLALSGATQTRMLTGFASRDKQALEMVGQITNEALSNIRTVAGIGKERRFI EALETELEKPFKTAIQKANIYGFCFAFAQCIMFIANSASYRYGGYLISNEGLHFSYVFRV ISAVVLSATALGRAFSYTPSYAKAKISAARFFQLLDRQPPISVYNTAGEKWDNFQGKIDF VDCKFTYPSRPDSQVLNGLSVSISPGQTLAFVGSSGCGKSTSIQLLERFYDPDQGKVMID GHDSKKVNVQFLRSNIGIVSQEPVLFACSIMDNIKYGDNTKEIPMERVIAAAKQAQLHDF VMSLPEKYETNVGSQGSQLSRGEKQRIAIARAIVRDPKILLLDEATSALDTESEKTVQVA LDKAREGRTCIVIAHRLSTIQNADIIAVMAQGVVIEKGTHEELMAQKGAYYKLVTTGSPI S Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| ADReCS ID | BADD_A06414 ; BADD_A08478 | |||||
| HIT2.0 ID | T64LKA | |||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Taurocholic acid | Ligand Info | |||||
| Structure Description | Human bile salt exporter ABCB11 in complex with taurocholate | PDB:7E1A | ||||
| Method | Electron microscopy | Resolution | 3.66 Å | Mutation | No | [2] |
| PDB Sequence |
GFFQLFRFSS
55 STDIWLMFVG65 SLCAFLHGIA75 QPGVLLIFGT85 MTDVFIDYDV95 ELQELQIPGK 105 ACVNNTIVWT115 NSSLNQNMTN125 GTRCGLLNIE135 SEMIKFASYY145 AGIAVAVLIT 155 GYIQICFWVI165 AAARQIQKMR175 KFYFRRIMRM185 EIGWFDCNSV195 GELNTRFSDD 205 INKINDAIAD215 QMALFIQRMT225 STICGFLLGF235 FRGWKLTLVI245 ISVSPLIGIG 255 AATIGLSVSK265 FTDYELKAYA275 KAGVVADEVI285 SSMRTVAAFG295 GEKREVERYE 305 KNLVFAQRWG315 IRKGIVMGFF325 TGFVWCLIFL335 CYALAFWYGS345 TLVLDEGEYT 355 PGTLVQIFLS365 VIVGALNLGN375 ASPCLEAFAT385 GRAAATSIFE395 TIDRKPIIDC 405 MSEDGYKLDR415 IKGEIEFHNV425 TFHYPSRPEV435 KILNDLNMVI445 KPGEMTALVG 455 PSGAGKSTAL465 QLIQRFYDPC475 EGMVTVDGHD485 IRSLNIQWLR495 DQIGIVEQEP 505 VLFSTTIAEN515 IRYGREDATM525 EDIVQAAKEA535 NAYNFIMDLP545 QQFDTLVGEG 555 GGQMSGGQKQ565 RVAIARALIR575 NPKILLLDMA585 TSALDNESEA595 MVQEVLSKIQ 605 HGHTIISVAH615 RLSTVRAADT625 IIGFEHGTAV635 ERGTHEELLE645 RKGVYFTLVT 655 LQSQVRRILK746 FSAPEWPYML756 VGSVGAAVNG766 TVTPLYAFLF776 SQILGTFSIP 786 DKEEQRSQIN796 GVCLLFVAMG806 CVSLFTQFLQ816 GYAFAKSGEL826 LTKRLRKFGF 836 RAMLGQDIAW846 FDDLRNSPGA856 LTTRLATDAS866 QVQGAAGSQI876 GMIVNSFTNV 886 TVAMIIAFSF896 SWKLSLVILC906 FFPFLALSGA916 TQTRMLTGFA926 SRDKQALEMV 936 GQITNEALSN946 IRTVAGIGKE956 RRFIEALETE966 LEKPFKTAIQ976 KANIYGFCFA 986 FAQCIMFIAN996 SASYRYGGYL1006 ISNEGLHFSY1016 VFRVISAVVL1026 SATALGRAFS 1036 YTPSYAKAKI1046 SAARFFQLLD1056 RQPPISVYNT1066 AGEKWDNFQG1076 KIDFVDCKFT 1086 YPSRPDSQVL1096 NGLSVSISPG1106 QTLAFVGSSG1116 CGKSTSIQLL1126 ERFYDPDQGK 1136 VMIDGHDSKK1146 VNVQFLRSNI1156 GIVSQEPVLF1166 ACSIMDNIKY1176 GDNTKEIPME 1186 RVIAAAKQAQ1196 LHDFVMSLPE1206 KYETNVGSQG1216 SQLSRGEKQR1226 IAIARAIVRD 1236 PKILLLDEAT1246 SALDTESEKT1256 VQVALDKARE1266 GRTCIVIAHR1276 LSTIQNADII 1286 AVMAQGVVIE1296 KGTHEELMAQ1306 KGAYYKLVT
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| ABC transporters | hsa02010 | Affiliated Target |
|
| Class: Environmental Information Processing => Membrane transport | Pathway Hierarchy | ||
| Bile secretion | hsa04976 | Affiliated Target |
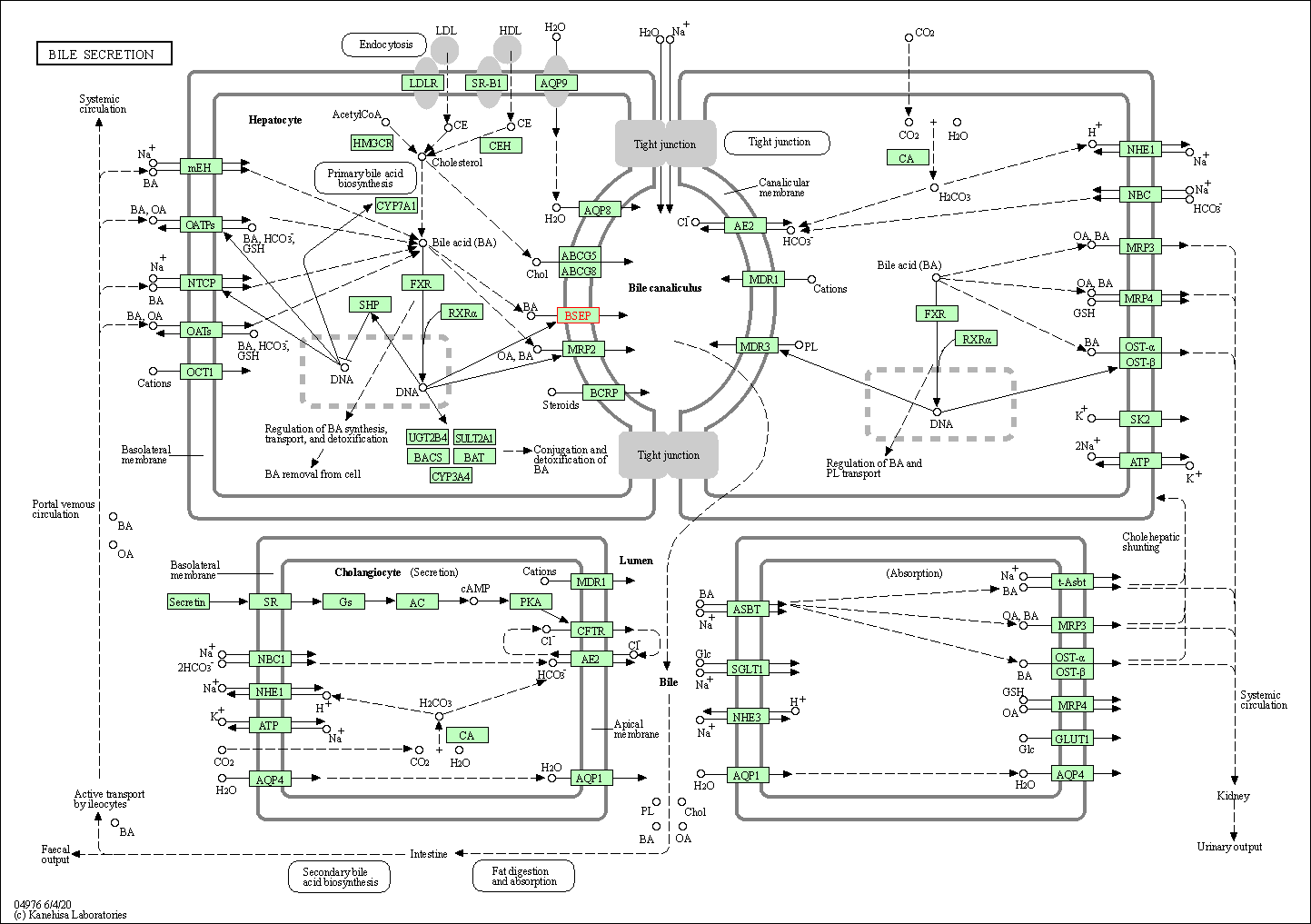
|
| Class: Organismal Systems => Digestive system | Pathway Hierarchy | ||
| Cholesterol metabolism | hsa04979 | Affiliated Target |
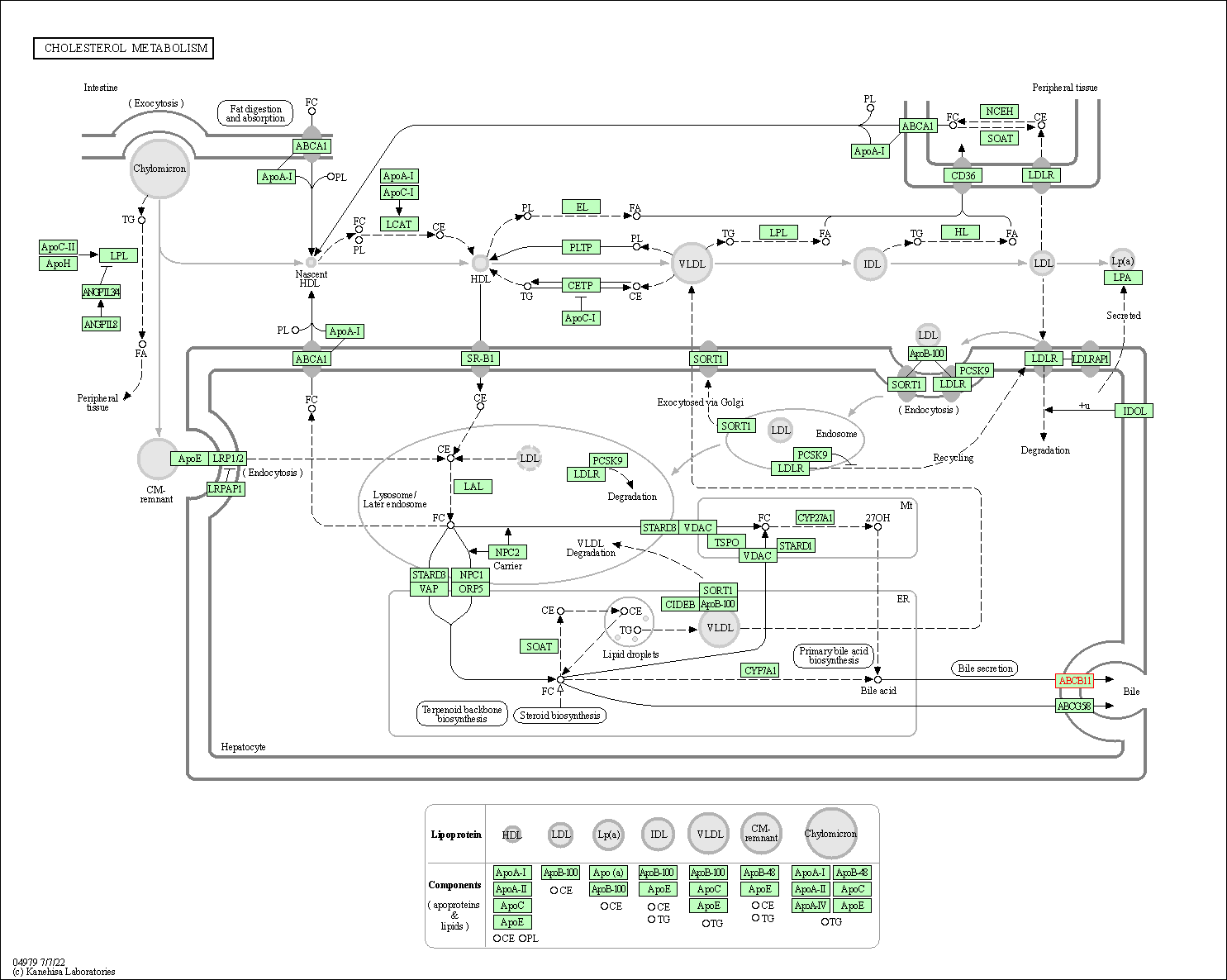
|
| Class: Organismal Systems => Digestive system | Pathway Hierarchy | ||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 2 KEGG Pathways | + | ||||
| 1 | ABC transporters | |||||
| 2 | Bile secretion | |||||
| Reactome | [+] 1 Reactome Pathways | + | ||||
| 1 | Recycling of bile acids and salts | |||||
| WikiPathways | [+] 5 WikiPathways | + | ||||
| 1 | Nuclear Receptors in Lipid Metabolism and Toxicity | |||||
| 2 | Nuclear Receptors Meta-Pathway | |||||
| 3 | Farnesoid X Receptor Pathway | |||||
| 4 | Drug Induction of Bile Acid Pathway | |||||
| 5 | Bile acid and bile salt metabolism | |||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013 Jan;41(Database issue):D991-5. | |||||
| REF 2 | Structures of human bile acid exporter ABCB11 reveal a transport mechanism facilitated by two tandem substrate-binding pockets. Cell Res. 2022 May;32(5):501-504. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

