Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T21782
(Former ID: TTDI00605)
|
|||||
| Target Name |
Proliferating cell nuclear antigen (PCNA)
|
|||||
| Synonyms |
Cyclin
Click to Show/Hide
|
|||||
| Gene Name |
PCNA
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Obesity [ICD-11: 5B80-5B81] | |||||
| Function |
Induces a robust stimulatory effect on the 3'-5' exonuclease and 3'-phosphodiesterase, but not apurinic-apyrimidinic (AP) endonuclease, APEX2 activities. Has to be loaded onto DNA in order to be able to stimulate APEX2. Plays a key role in DNA damage response (DDR) by being conveniently positioned at the replication fork to coordinate DNA replication with DNA repair and DNA damage tolerance pathways. Acts as a loading platform to recruit DDR proteins that allow completion of DNA replication after DNA damage and promote postreplication repair: Monoubiquitinated PCNA leads to recruitment of translesion (TLS) polymerases, while 'Lys-63'-linked polyubiquitination of PCNA is involved in error-free pathway and employs recombination mechanisms to synthesize across the lesion. Auxiliary protein of DNA polymerase delta and is involved in the control of eukaryotic DNA replication by increasing the polymerase's processibility during elongation of the leading strand.
Click to Show/Hide
|
|||||
| UniProt ID | ||||||
| Sequence |
MFEARLVQGSILKKVLEALKDLINEACWDISSSGVNLQSMDSSHVSLVQLTLRSEGFDTY
RCDRNLAMGVNLTSMSKILKCAGNEDIITLRAEDNADTLALVFEAPNQEKVSDYEMKLMD LDVEQLGIPEQEYSCVVKMPSGEFARICRDLSHIGDAVVISCAKDGVKFSASGELGNGNI KLSQTSNVDKEEEAVTIEMNEPVQLTFALRYLNFFTKATPLSSTVTLSMSADVPLVVEYK IADMGHLKYYLAPKIEDEEGS Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| ADReCS ID | BADD_A08298 | |||||
| HIT2.0 ID | T09KQF | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 1 Clinical Trial Drugs | + | ||||
| 1 | ATX-101 | Drug Info | Phase 3 | Obesity | [1], [2] | |
| Discontinued Drug(s) | [+] 1 Discontinued Drugs | + | ||||
| 1 | ChelASE | Drug Info | Discontinued in Phase 1 | Retina disorder | [3] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Immunomodulator | [+] 1 Immunomodulator drugs | + | ||||
| 1 | ATX-101 | Drug Info | [1] | |||
| Inhibitor | [+] 1 Inhibitor drugs | + | ||||
| 1 | ChelASE | Drug Info | [1], [4] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Liothyronine | Ligand Info | |||||
| Structure Description | Structure of PCNA | PDB:3VKX | ||||
| Method | X-ray diffraction | Resolution | 2.10 Å | Mutation | No | [5] |
| PDB Sequence |
MFEARLVQGS
10 ILKKVLEALK20 DLINEACWDI30 SSSGVNLQSM40 DSSHVSLVQL50 TLRSEGFDTY 60 RCDRNLAMGV70 NLTSMSKILK80 CAGNEDIITL90 RAEDNADTLA100 LVFEAPNQEK 110 VSDYEMKLMD120 LDVEQLGIPE130 QEYSCVVKMP140 SGEFARICRD150 LSHIGDAVVI 160 SCAKDGVKFS170 ASGELGNGNI180 KLSQTEAVTI197 EMNEPVQLTF207 ALRYLNFFTK 217 ATPLSSTVTL227 SMSADVPLVV237 EYKIADMGHL247 KYYLAPKI
|
|||||
|
|
||||||
| Ligand Name: 4-[4-[(2S)-2-amino-3-hydroxypropyl]-2,6-diiodophenoxy]phenol | Ligand Info | |||||
| Structure Description | Structure of PCNA bound to a small molecule inhibitor | PDB:3WGW | ||||
| Method | X-ray diffraction | Resolution | 2.80 Å | Mutation | No | [6] |
| PDB Sequence |
MFEARLVQGS
10 ILKKVLEALK20 DLINEACWDI30 SSSGVNLQSM40 DSSHVSLVQL50 TLRSEGFDTY 60 RCDRNLAMGV70 NLTSMSKILK80 CAGNEDIITL90 RAEDNADTLA100 LVFEAPNQEK 110 VSDYEMKLMD120 LDVEQLGIPE130 QEYSCVVKMP140 SGEFARICRD150 LSHIGDAVVI 160 SCAKDGVKFS170 ASGELGNGNI180 KLSQTEAVTI197 EMNEPVQLTF207 ALRYLNFFTK 217 ATPLSSTVTL227 SMSADVPLVV237 EYKIADMGHL247 KYYLAPKI
|
|||||
|
|
MET40
3.381
HIS44
3.505
VAL45
3.961
SER46
4.632
LEU47
3.548
GLU124
3.030
LEU126
3.968
GLY127
3.069
ILE128
3.977
PRO129
3.819
GLN131
4.003
TYR133
4.423
|
|||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
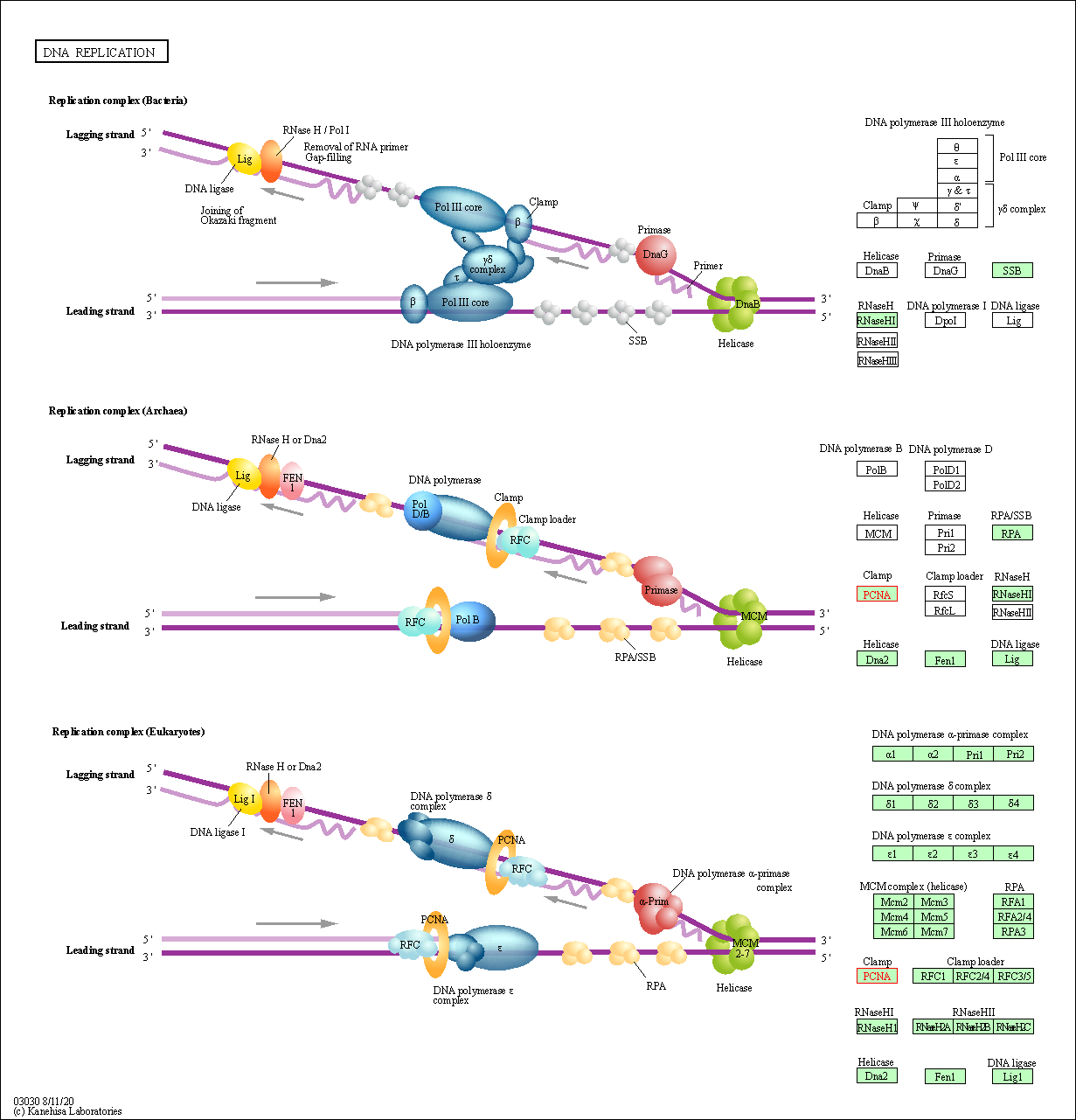
| DNA replication | hsa03030 | Affiliated Target |

|
| Class: Genetic Information Processing => Replication and repair | Pathway Hierarchy | ||
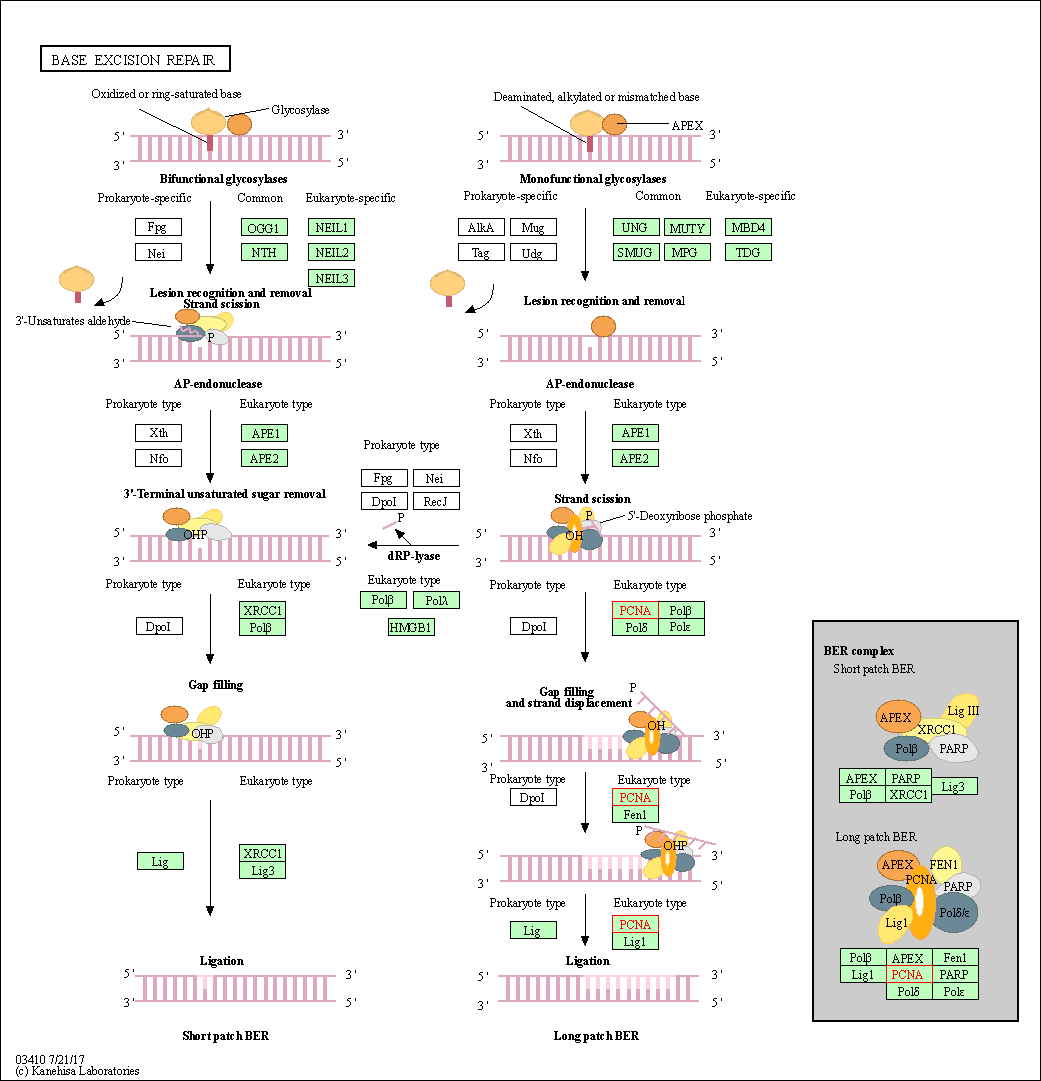
| Base excision repair | hsa03410 | Affiliated Target |

|
| Class: Genetic Information Processing => Replication and repair | Pathway Hierarchy | ||
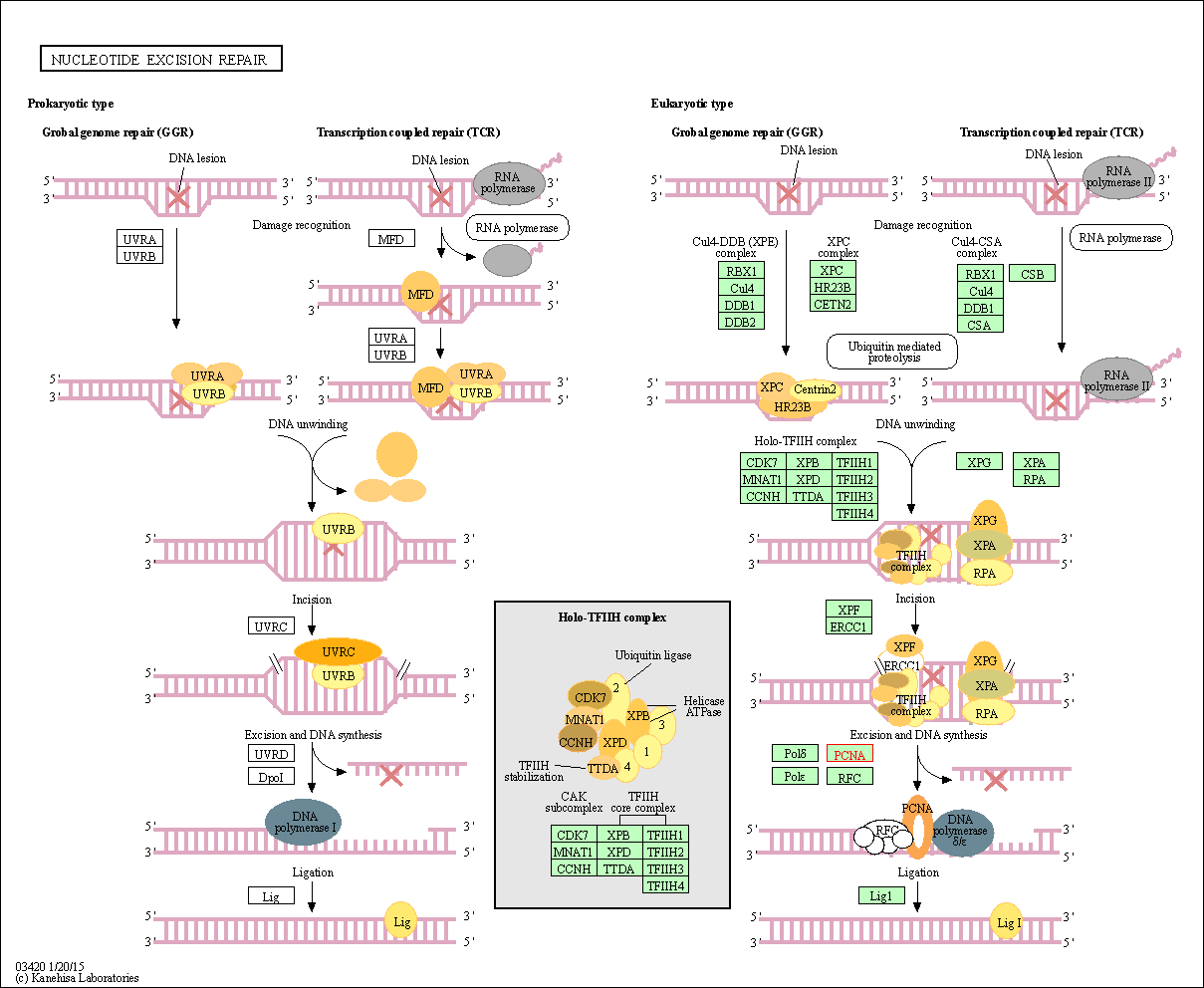
| Nucleotide excision repair | hsa03420 | Affiliated Target |

|
| Class: Genetic Information Processing => Replication and repair | Pathway Hierarchy | ||
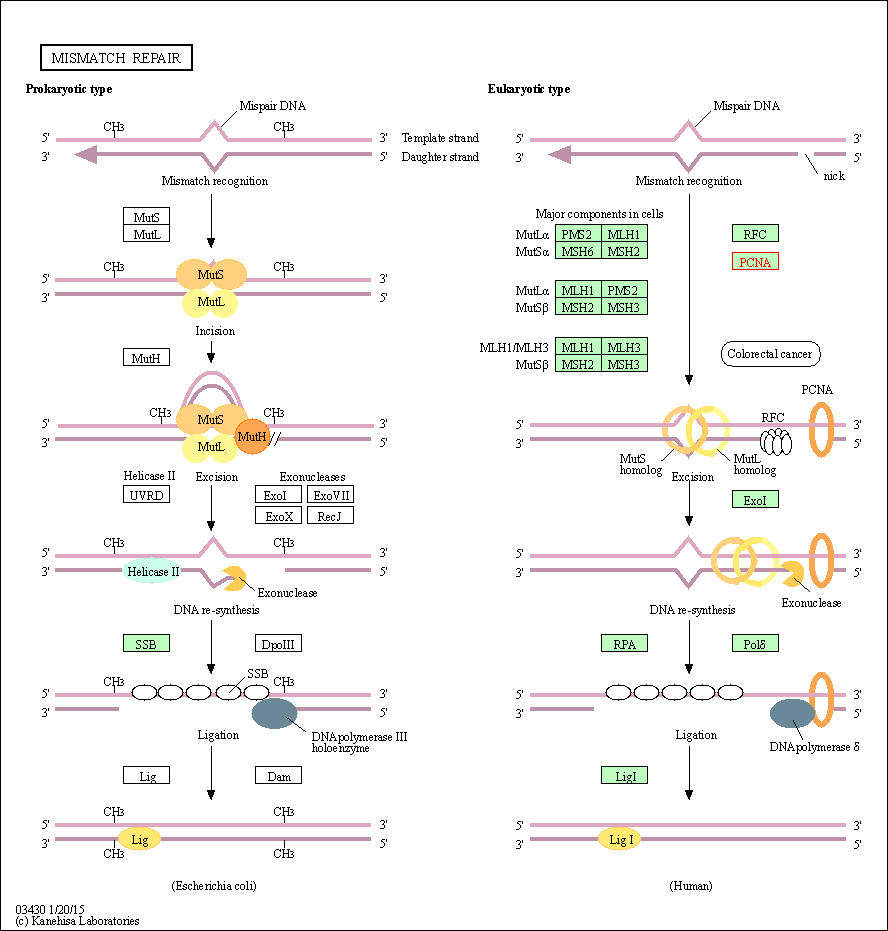
| Mismatch repair | hsa03430 | Affiliated Target |

|
| Class: Genetic Information Processing => Replication and repair | Pathway Hierarchy | ||
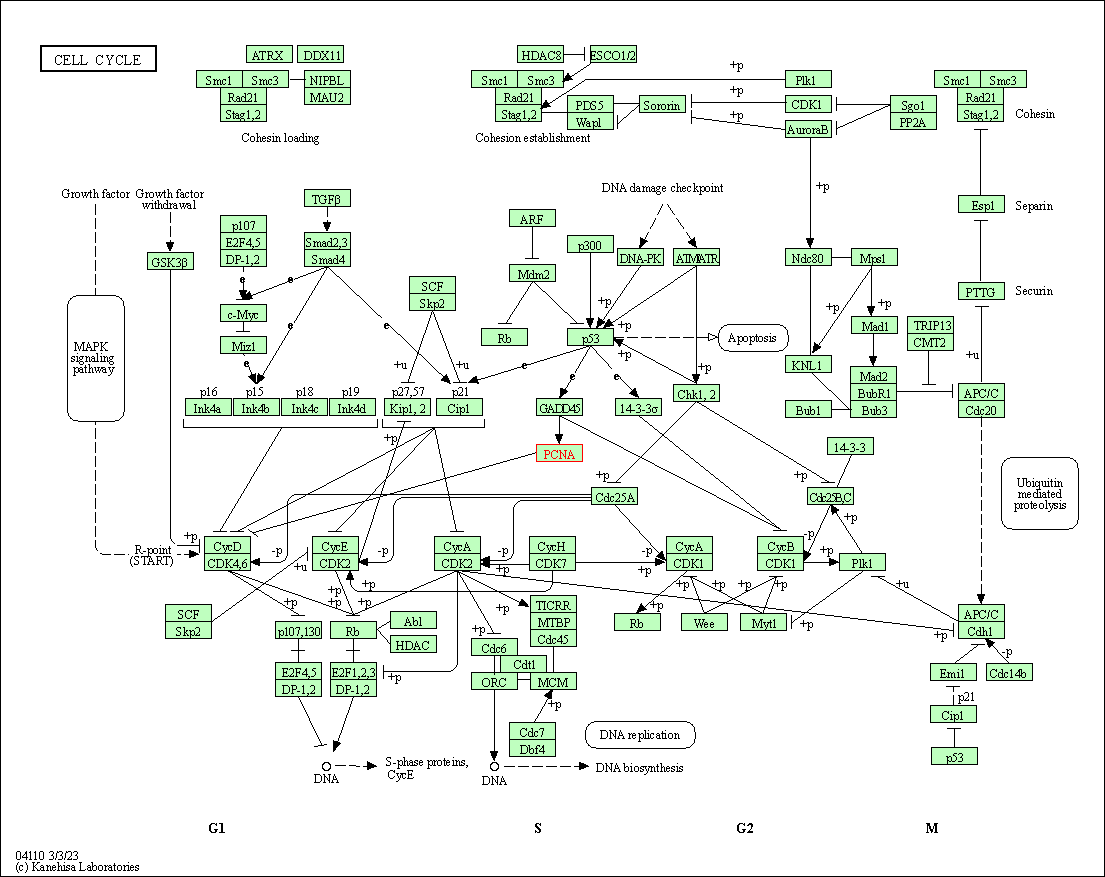
| Cell cycle | hsa04110 | Affiliated Target |

|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
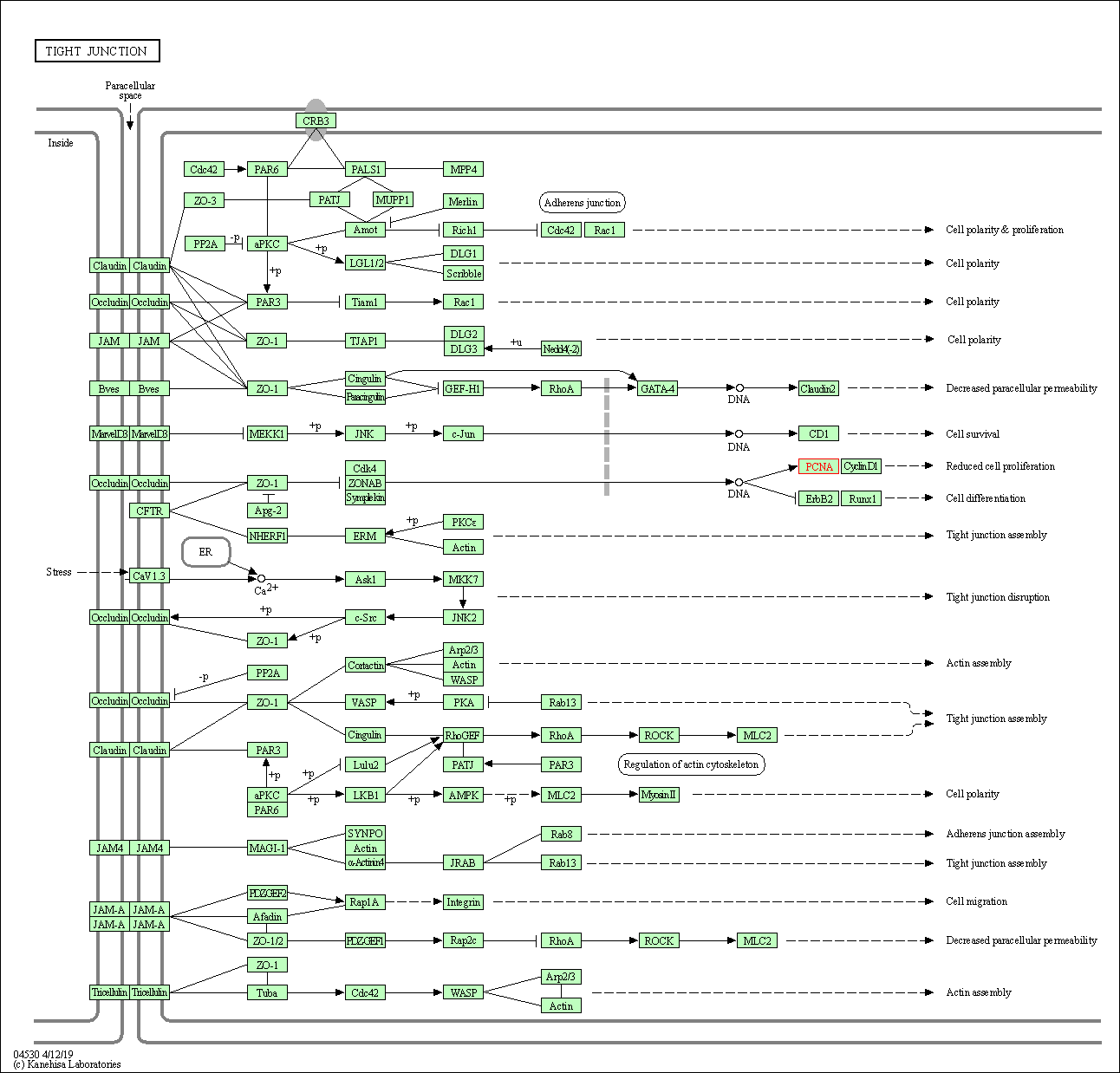
| Tight junction | hsa04530 | Affiliated Target |

|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 81 | Degree centrality | 8.70E-03 | Betweenness centrality | 5.27E-03 |
|---|---|---|---|---|---|
| Closeness centrality | 2.45E-01 | Radiality | 1.43E+01 | Clustering coefficient | 1.28E-01 |
| Neighborhood connectivity | 3.47E+01 | Topological coefficient | 4.20E-02 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating Transcription Factors | ||||||
| Target-interacting Proteins | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Targeting proliferating cell nuclear antigen and its protein interactions induces apoptosis in multiple myeloma cells. PLoS One. 2013 Jul 31;8(7):e70430. | |||||
| REF 2 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 610). | |||||
| REF 3 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800017704) | |||||
| REF 4 | Safety and efficacy assessment of chimeric ribozyme to proliferating cell nuclear antigen to prevent recurrence of proliferative vitreoretinopathy. Arch Ophthalmol. 2007 Sep;125(9):1161-7. | |||||
| REF 5 | Identification of small molecule proliferating cell nuclear antigen (PCNA) inhibitor that disrupts interactions with PIP-box proteins and inhibits DNA replication. J Biol Chem. 2012 Apr 20;287(17):14289-300. | |||||
| REF 6 | A small molecule inhibitor of monoubiquitinated Proliferating Cell Nuclear Antigen (PCNA) inhibits repair of interstrand DNA cross-link, enhances DNA double strand break, and sensitizes cancer cells to cisplatin. J Biol Chem. 2014 Mar 7;289(10):7109-7120. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

