Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T21705
(Former ID: TTDR00937)
|
|||||
| Target Name |
F-box and WD-40 domain protein 7 (Fbxw7)
|
|||||
| Synonyms |
hCdc4; hAgo; SEL10; SEL-10; FBX30; FBW7; F-box/WD repeat-containing protein 7; F-box-WD40 repeat protein 6; F-box protein Fbxw6; F-box protein FBX30; F-box protein FBW7; F-box and WD-40 domain-containing protein 7; Archipelago homolog
Click to Show/Hide
|
|||||
| Gene Name |
FBXW7
|
|||||
| Target Type |
Literature-reported target
|
[1] | ||||
| Function |
Recognizes and binds phosphorylated sites/phosphodegrons within target proteins and thereafter bring them to the SCF complex for ubiquitination. Identified substrates include cyclin-E (CCNE1 or CCNE2), DISC1, JUN, MYC, NOTCH1 released notch intracellular domain (NICD), NOTCH2, MCL1, and probably PSEN1. Acts as a negative regulator of JNK signaling by binding to phosphorylated JUN and promoting its ubiquitination and subsequent degradation. SCF(FBXW7) complex mediates the ubiquitination and subsequent degradation of NFE2L1. Involved in bone homeostasis and negative regulation of osteoclast differentiation. Regulates the amplitude of the cyclic expression of hepatic core clock genes and genes involved in lipid and glucose metabolism via ubiquitination and proteasomal degradation of their transcriptional repressor NR1D1; CDK1-dependent phosphorylation of NR1D1 is necessary for SCF(FBXW7)-mediated ubiquitination. Substrate recognition component of a SCF (SKP1-CUL1-F-box protein) E3 ubiquitin-protein ligase complex which mediates the ubiquitination and subsequent proteasomal degradation of target proteins.
Click to Show/Hide
|
|||||
| UniProt ID | ||||||
| Sequence |
MNQELLSVGSKRRRTGGSLRGNPSSSQVDEEQMNRVVEEEQQQQLRQQEEEHTARNGEVV
GVEPRPGGQNDSQQGQLEENNNRFISVDEDSSGNQEEQEEDEEHAGEQDEEDEEEEEMDQ ESDDFDQSDDSSREDEHTHTNSVTNSSSIVDLPVHQLSSPFYTKTTKMKRKLDHGSEVRS FSLGKKPCKVSEYTSTTGLVPCSATPTTFGDLRAANGQGQQRRRITSVQPPTGLQEWLKM FQSWSGPEKLLALDELIDSCEPTQVKHMMQVIEPQFQRDFISLLPKELALYVLSFLEPKD LLQAAQTCRYWRILAEDNLLWREKCKEEGIDEPLHIKRRKVIKPGFIHSPWKSAYIRQHR IDTNWRRGELKSPKVLKGHDDHVITCLQFCGNRIVSGSDDNTLKVWSAVTGKCLRTLVGH TGGVWSSQMRDNIIISGSTDRTLKVWNAETGECIHTLYGHTSTVRCMHLHEKRVVSGSRD ATLRVWDIETGQCLHVLMGHVAAVRCVQYDGRRVVSGAYDFMVKVWDPETETCLHTLQGH TNRVYSLQFDGIHVVSGSLDTSIRVWDVETGNCIHTLTGHQSLTSGMELKDNILVSGNAD STVKIWDIKTGQCLQTLQGPNKHQSAVTCLQFNKNFVITSSDDGTVKLWDLKTGEFIRNL VTLESGGSGGVVWRIRASNTKLVCAVGSRNGTEETKLLVLDFDVDMK Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| HIT2.0 ID | T01N30 | |||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Pathway Affiliation
Biological Network Descriptors
|
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Ubiquitin mediated proteolysis | hsa04120 | Affiliated Target |
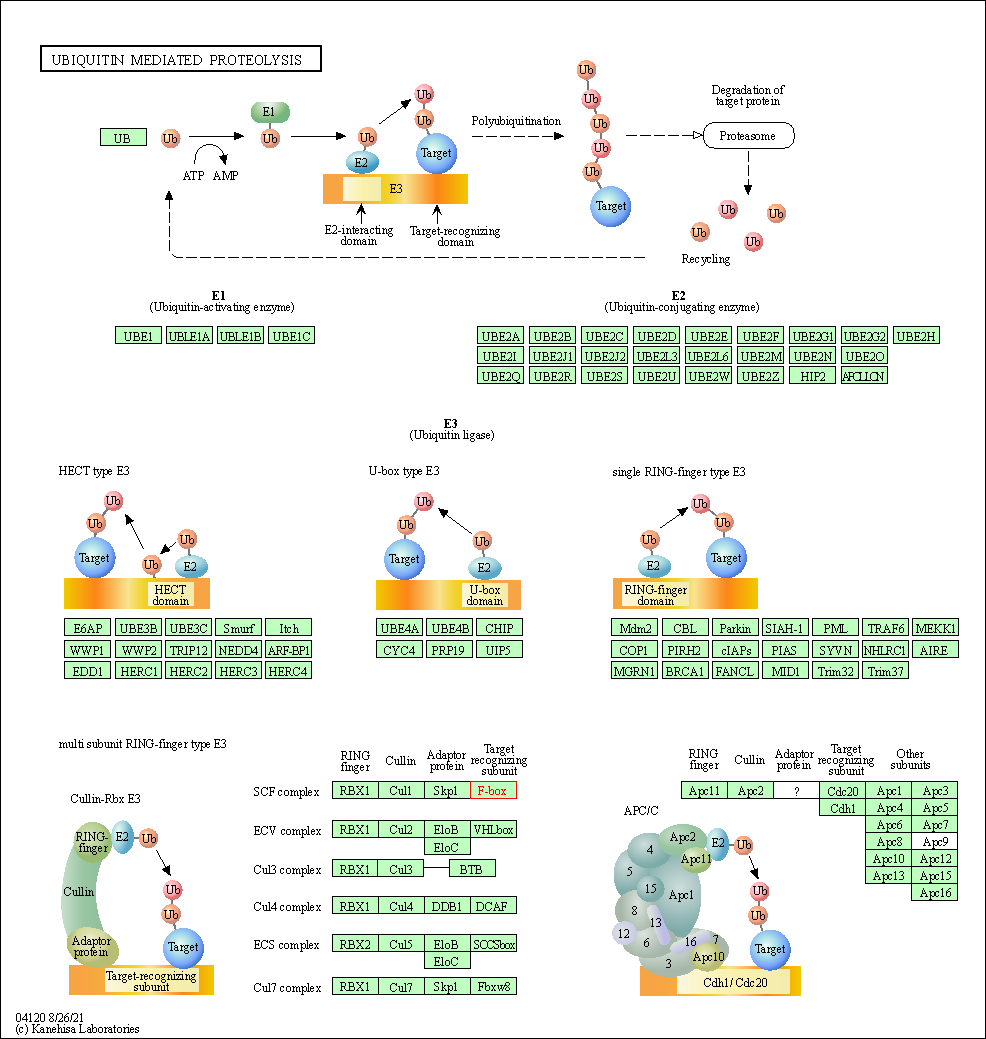
|
| Class: Genetic Information Processing => Folding, sorting and degradation | Pathway Hierarchy | ||
| Degree | 16 | Degree centrality | 1.72E-03 | Betweenness centrality | 5.58E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 2.38E-01 | Radiality | 1.42E+01 | Clustering coefficient | 1.42E-01 |
| Neighborhood connectivity | 4.93E+01 | Topological coefficient | 8.73E-02 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | BRAF and FBXW7 (CDC4, FBW7, AGO, SEL10) mutations in distinct subsets of pancreatic cancer: potential therapeutic targets. Am J Pathol. 2003 Oct;163(4):1255-60. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

