Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T21689
(Former ID: TTDR00411)
|
|||||
| Target Name |
Apolipoprotein E (APOE)
|
|||||
| Synonyms |
ApoE; Apo-E
Click to Show/Hide
|
|||||
| Gene Name |
APOE
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 3 Target-related Diseases | + | ||||
| 1 | Hyper-lipoproteinaemia [ICD-11: 5C80] | |||||
| 2 | Intracerebral haemorrhage [ICD-11: 8B00] | |||||
| 3 | Alzheimer disease [ICD-11: 8A20] | |||||
| Function |
APOE is a core component of plasma lipoproteins and is involved in their production, conversion and clearance. Apoliproteins are amphipathic molecules that interact both with lipids of the lipoprotein particle core and the aqueous environment of the plasma. As such, APOE associates with chylomicrons, chylomicron remnants, very low density lipoproteins (VLDL) and intermediate density lipoproteins (IDL) but shows a preferential binding to high-density lipoproteins (HDL). It also binds a wide range of cellular receptors including the LDL receptor/LDLR, the LDL receptor-related proteins LRP1, LRP2 and LRP8 and the very low-density lipoprotein receptor/VLDLR that mediate the cellular uptake of the APOE-containing lipoprotein particles. Finally, APOE has also a heparin-binding activity and binds heparan-sulfate proteoglycans on the surface of cells, a property that supports the capture and the receptor-mediated uptake of APOE-containing lipoproteins by cells. A main function of APOE is to mediate lipoprotein clearance through the uptake of chylomicrons, VLDLs, and HDLs by hepatocytes. APOE is also involved in the biosynthesis by the liver of VLDLs as well as their uptake by peripheral tissues ensuring the delivery of triglycerides and energy storage in muscle, heart and adipose tissues. By participating to the lipoprotein-mediated distribution of lipids among tissues, APOE plays a critical role in plasma and tissues lipid homeostasis. APOE is also involved in two steps of reverse cholesterol transport, the HDLs-mediated transport of cholesterol from peripheral tissues to the liver, and thereby plays an important role in cholesterol homeostasis. First, it is functionally associated with ABCA1 in the biogenesis of HDLs in tissues. Second, it is enriched in circulating HDLs and mediates their uptake by hepatocytes. APOE also plays an important role in lipid transport in the central nervous system, regulating neuron survival and sprouting. APOE in also involved in innate and adaptive immune responses, controlling for instance the survival of myeloid-derived suppressor cells. APOE, may also play a role in transcription regulation through a receptor-dependent and cholesterol-independent mechanism, that activates MAP3K12 and a non-canonical MAPK signal transduction pathway that results in enhanced AP-1-mediated transcription of APP. APOE is an apolipoprotein, a protein associating with lipid particles, that mainly functions in lipoprotein-mediated lipid transport between organs via the plasma and interstitial fluids.
Click to Show/Hide
|
|||||
| BioChemical Class |
Apolipoprotein
|
|||||
| UniProt ID | ||||||
| Sequence |
MKVLWAALLVTFLAGCQAKVEQAVETEPEPELRQQTEWQSGQRWELALGRFWDYLRWVQT
LSEQVQEELLSSQVTQELRALMDETMKELKAYKSELEEQLTPVAEETRARLSKELQAAQA RLGADMEDVCGRLVQYRGEVQAMLGQSTEELRVRLASHLRKLRKRLLRDADDLQKRLAVY QAGAREGAERGLSAIRERLGPLVEQGRVRAATVGSLAGQPLQERAQAWGERLRARMEEMG SRTRDRLDEVKEQVAEVRAKLEEQAQQIRLQAEAFQARLKSWFEPLVEDMQRQWAGLVEK VQAAVGTSAAPVPSDNH Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| ADReCS ID | BADD_A01187 ; BADD_A02936 | |||||
| HIT2.0 ID | T97ESI | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 3 Clinical Trial Drugs | + | ||||
| 1 | AEM-28 | Drug Info | Phase 2 | Familial hypercholesterolemia | [2] | |
| 2 | CN-105 | Drug Info | Phase 2 | Nontraumatic intracerebral hemorrhage | [2], [3], [4] | |
| 3 | LX1001 | Drug Info | Phase 1/2 | Alzheimer disease | [5] | |
| Discontinued Drug(s) | [+] 1 Discontinued Drugs | + | ||||
| 1 | Apolipoprotein E | Drug Info | Terminated | Glaucoma/ocular hypertension | [6] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Modulator | [+] 2 Modulator drugs | + | ||||
| 1 | AEM-28 | Drug Info | [1] | |||
| 2 | Apolipoprotein E | Drug Info | [8] | |||
| Agonist | [+] 1 Agonist drugs | + | ||||
| 1 | CN-105 | Drug Info | [2], [3], [4] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: 1-[5-Chloro-4'-(2-hydroxypropan-2-yl)[1,1'-biphenyl]-3-yl]cyclobutane-1-carboximidamide | Ligand Info | |||||
| Structure Description | Fragment-based Discovery of an apoE4 Stabilizer | PDB:6NCO | ||||
| Method | X-ray diffraction | Resolution | 1.71 Å | Mutation | Yes | [9] |
| PDB Sequence |
QRWELALGRF
33 WDYLRWVQTL43 SEQVQEELLS53 SQVTQELRAL63 MDETMKELKA73 YKSELEEQLT 83 PVAEETRARL93 SKELQAAQAR103 LGADMEDVRG113 RLVQYRGEVQ123 AMLGQSTEEL 133 RVRLASHLRK143 LRKRLLRDAD153 DLQKRLAVYQ163 A
|
|||||
|
|
||||||
| Ligand Name: 1-(3-Chlorophenyl)cyclobutane-1-carboximidamide | Ligand Info | |||||
| Structure Description | Fragment-based Discovery of an apoE4 Stabilizer | PDB:6NCN | ||||
| Method | X-ray diffraction | Resolution | 1.82 Å | Mutation | Yes | [9] |
| PDB Sequence |
QRWELALGRF
33 WDYLRWVQTL43 SEQVQEELLS53 SQVTQELRAL63 MDETMKELKA73 YKSELEEQLT 83 PVAEETRARL93 SKELQAAQAR103 LGADMEDVRG113 RLVQYRGEVQ123 AMLGQSTEEL 133 RVRLASHLRK143 LRKRLLRDAD153 DLQKRLAVYQ163 A
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Cholesterol metabolism | hsa04979 | Affiliated Target |
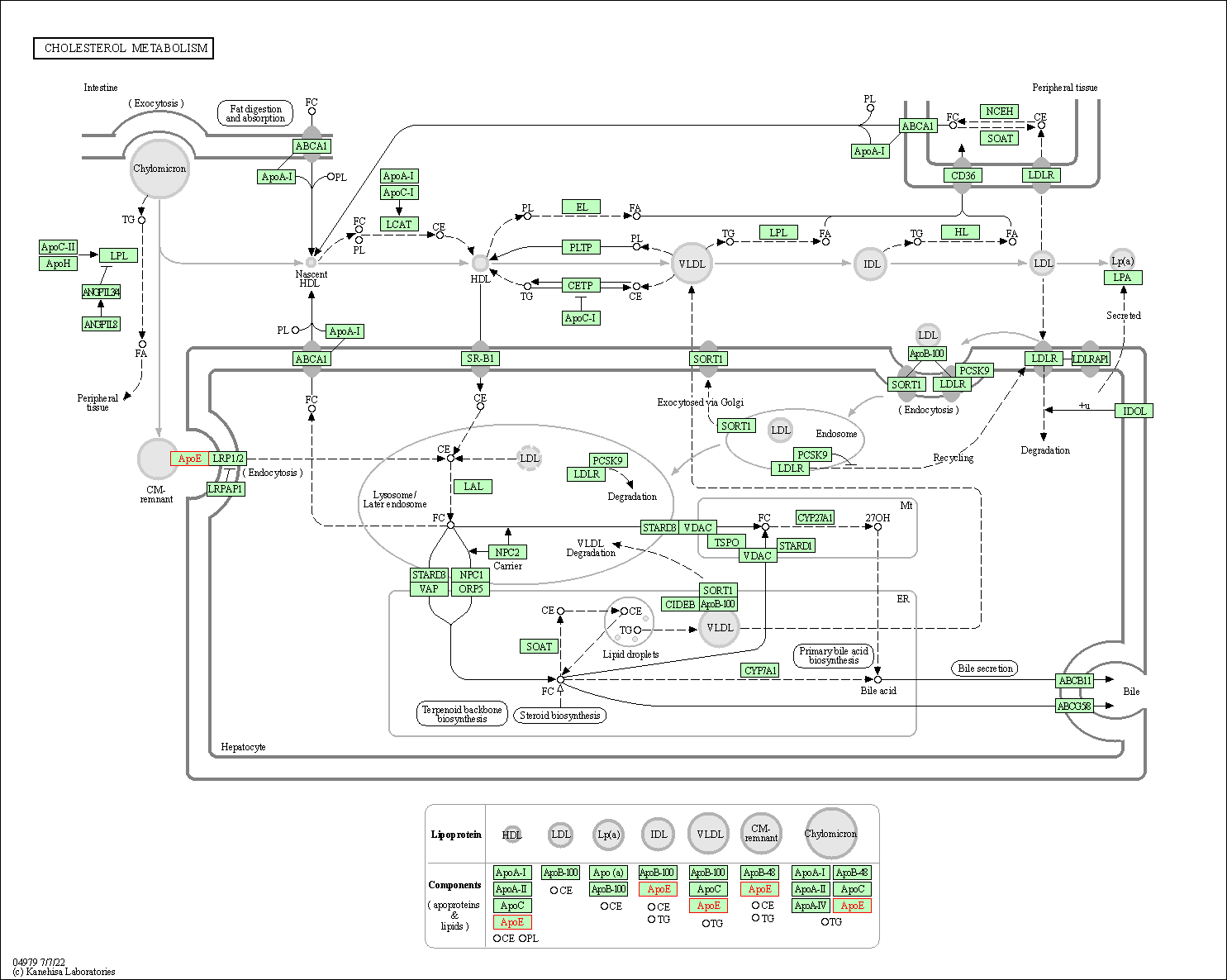
|
| Class: Organismal Systems => Digestive system | Pathway Hierarchy | ||
| Degree | 32 | Degree centrality | 3.44E-03 | Betweenness centrality | 3.20E-03 |
|---|---|---|---|---|---|
| Closeness centrality | 2.30E-01 | Radiality | 1.40E+01 | Clustering coefficient | 1.27E-01 |
| Neighborhood connectivity | 1.52E+01 | Topological coefficient | 5.41E-02 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-regulating Transcription Factors | ||||||
| Target-interacting Proteins | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 1 KEGG Pathways | + | ||||
| 1 | Alzheimer's disease | |||||
| NetPath Pathway | [+] 3 NetPath Pathways | + | ||||
| 1 | TGF_beta_Receptor Signaling Pathway | |||||
| 2 | FSH Signaling Pathway | |||||
| 3 | Wnt Signaling Pathway | |||||
| Reactome | [+] 4 Reactome Pathways | + | ||||
| 1 | Chylomicron-mediated lipid transport | |||||
| 2 | HDL-mediated lipid transport | |||||
| 3 | Scavenging by Class A Receptors | |||||
| 4 | Retinoid metabolism and transport | |||||
| WikiPathways | [+] 7 WikiPathways | + | ||||
| 1 | Statin Pathway | |||||
| 2 | Apoptosis-related network due to altered Notch3 in ovarian cancer | |||||
| 3 | Binding and Uptake of Ligands by Scavenger Receptors | |||||
| 4 | Visual phototransduction | |||||
| 5 | Lipid digestion, mobilization, and transport | |||||
| 6 | Alzheimers Disease | |||||
| 7 | Vitamin B12 Metabolism | |||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Clinical pipeline report, company report or official report of LipimetiX. | |||||
| REF 2 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 3 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 4 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 5 | ClinicalTrials.gov (NCT03634007) A 52-Week, Multicenter, Phase 1/2 Open-label Study to Evaluate the Safety of LX1001 in Participants With APOE4 Homozygote Alzheimer's Disease. U.S.National Institutes of Health. | |||||
| REF 6 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800000261) | |||||
| REF 7 | Clinical pipeline report, company report or official report of Lexeo Therapeutics | |||||
| REF 8 | Apolipoprotein E: a potent inhibitor of endothelial and tumor cell proliferation. J Cell Biochem. 1994 Mar;54(3):299-308. | |||||
| REF 9 | Fragment-Based Discovery of an Apolipoprotein E4 (apoE4) Stabilizer. J Med Chem. 2019 Apr 25;62(8):4120-4130. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

