Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T11843
(Former ID: TTDS00398)
|
|||||
| Target Name |
Vitamin K epoxide reductase complex 1 (VKORC1)
|
|||||
| Synonyms |
Vitamin K1 2,3-epoxide reductase subunit 1; VKORC1; VKOR; UNQ308/PRO351; MSTP576; MSTP134
Click to Show/Hide
|
|||||
| Gene Name |
VKORC1
|
|||||
| Target Type |
Successful target
|
[1] | ||||
| Disease | [+] 5 Target-related Diseases | + | ||||
| 1 | Bleeding disorder [ICD-11: GA20-GA21] | |||||
| 2 | Coagulation defect [ICD-11: 3B10] | |||||
| 3 | Pulmonary thromboembolism [ICD-11: BB00] | |||||
| 4 | Supraventricular tachyarrhythmia [ICD-11: BC81] | |||||
| 5 | Thrombosis [ICD-11: DB61-GB90] | |||||
| Function |
Involved invitamin K metabolism. Catalytic subunit of the vitamin K epoxide reductase (VKOR) complex which reduces inactive vitamin K 2,3-epoxide to active vitamin K. Vitamin K is required for the gamma-carboxylation of various proteins, including clotting factors, and is required for normal blood coagulation, but also for normal bone development.
Click to Show/Hide
|
|||||
| BioChemical Class |
Short-chain dehydrogenases reductase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 1.17.4.4
|
|||||
| Sequence |
MGSTWGSPGWVRLALCLTGLVLSLYALHVKAARARDRDYRALCDVGTAISCSRVFSSRWG
RGFGLVEHVLGQDSILNQSNSIFGCIFYTLQLLLGCLRTRWASVLMLLSSLVSLAGSVYL AWILFFVLYDFCIVCITTYAINVSLMWLSFRKVQEPQGKAKRH Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Approved Drug(s) | [+] 5 Approved Drugs | + | ||||
| 1 | Acenocoumarol | Drug Info | Approved | Thrombosis | [2] | |
| 2 | Dicumarol | Drug Info | Approved | Thrombosis | [3], [4], [5] | |
| 3 | Phenindione | Drug Info | Approved | Coagulation defect | [5], [6], [7] | |
| 4 | Phenprocoumon | Drug Info | Approved | Thrombosis | [5], [8], [9] | |
| 5 | Warfarin | Drug Info | Approved | Atrial fibrillation | [10], [11] | |
| Clinical Trial Drug(s) | [+] 1 Clinical Trial Drugs | + | ||||
| 1 | Tecarfarin | Drug Info | Phase 3 | Cerebrovascular ischaemia | [12] | |
| Mode of Action | [+] 1 Modes of Action | + | ||||
| Inhibitor | [+] 6 Inhibitor drugs | + | ||||
| 1 | Acenocoumarol | Drug Info | [13], [14], [15] | |||
| 2 | Dicumarol | Drug Info | [1] | |||
| 3 | Phenindione | Drug Info | [15] | |||
| 4 | Phenprocoumon | Drug Info | [15] | |||
| 5 | Warfarin | Drug Info | [15] | |||
| 6 | Tecarfarin | Drug Info | [16] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Warfarin | Ligand Info | |||||
| Structure Description | Human VKOR with warfarin | PDB:6WV3 | ||||
| Method | X-ray diffraction | Resolution | 2.20 Å | Mutation | Yes | [17] |
| PDB Sequence |
KGEELFTGVV
12 PILVELDGDV22 NGHKFSVRGE32 GEGDATNGKL42 TLKFICTTGK52 LPVPWPTLVT 62 TLVQCFSRYP75 DHMKRHDFFK85 SAMPEGYVQE95 RTISFKDDGT105 YKTRAEVKFE 115 GDTLVNRIEL125 KGIDFKEDGN135 ILGHKLEYNS145 TWGSPGWVRL155 ALCLTGLVLS 165 LYALHVKAAR175 ARDRDYRALC185 DVGTAISCSR195 VFSSRWGRGF205 GLVEHVLGQD 215 SILNQSNSIF225 GCIFYTLQLL235 LGCLRTRWAS245 VLMLLSSLVS255 LAGSVYLAWI 265 LFFVLYDFCI275 VCITTYAINV285 SLMWLSFRKV295 QENSHNVYIT305 ADKQKNGIKA 315 NFKIRHNVED325 GSVQLADHYQ335 QNTPIGDGPV345 LLPDNHYLST355 QSVLSKDPNE 365 KRDHMVLLEF375 VTAAGITHHH385
|
|||||
|
|
LEU164
4.218
SER165
4.752
ALA168
3.667
VAL196
3.542
PHE197
3.701
SER199
3.864
TRP201
3.714
GLY202
3.730
PHE205
3.760
ASN222
2.762
SER223
3.938
|
|||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Ligand Name: Phenindione | Ligand Info | |||||
| Structure Description | Human VKOR with phenindione | PDB:6WV6 | ||||
| Method | X-ray diffraction | Resolution | 2.70 Å | Mutation | Yes | [17] |
| PDB Sequence |
SKGEELFTGV
11 VPILVELDGD21 VNGHKFSVRG31 EGEGDATNGK41 LTLKFICTTG51 KLPVPWPTLV 61 TTLVQCFSRY74 PDHMKRHDFF84 KSAMPEGYVQ94 ERTISFKDDG104 TYKTRAEVKF 114 EGDTLVNRIE124 LKGIDFKEDG134 NILGHKLEYN144 STWGSPGWVR154 LALCLTGLVL 164 SLYALHVKAA174 RARDRDYRAL184 CDVGTAISCS194 RVFSSRWGRG204 FGLVEHVLGQ 214 DSILNQSNSI224 FGCIFYTLQL234 LLGCLRTRWA244 SVLMLLSSLV254 SLAGSVYLAW 264 ILFFVLYDFC274 IVCITTYAIN284 VSLMWLSFRK294 VQENSHNVYI304 TADKQKNGIK 314 ANFKIRHNVE324 DGSVQLADHY334 QQNTPIGDGP344 VLLPDNHYLS354 TQSVLSKDPN 364 EKRDHMVLLE374 FVTAAGITHH384 H
|
|||||
|
|
LEU164
4.012
SER165
3.421
ALA168
3.682
VAL196
2.946
PHE197
2.643
SER199
3.701
TRP201
3.432
GLY202
3.449
ASN222
2.990
SER223
2.886
|
|||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
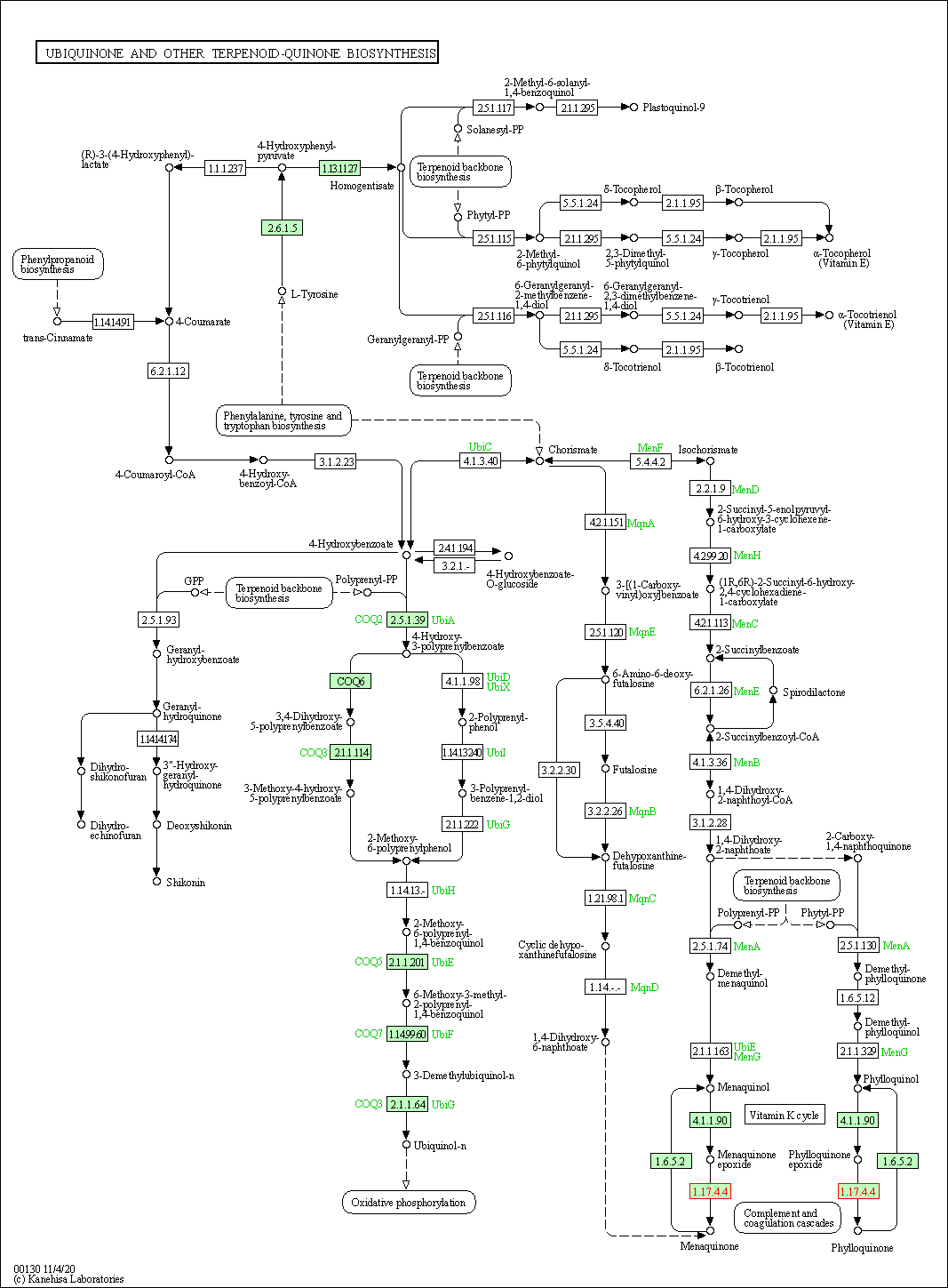
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Ubiquinone and other terpenoid-quinone biosynthesis | hsa00130 | Affiliated Target |

|
| Class: Metabolism => Metabolism of cofactors and vitamins | Pathway Hierarchy | ||
| Degree | 1 | Degree centrality | 1.07E-04 | Betweenness centrality | 0.00E+00 |
|---|---|---|---|---|---|
| Closeness centrality | 1.51E-01 | Radiality | 1.20E+01 | Clustering coefficient | 0.00E+00 |
| Neighborhood connectivity | 9.00E+00 | Topological coefficient | 1.00E+00 | Eccentricity | 14 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Drug Resistance Mutation (DRM) |
||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 1 KEGG Pathways | + | ||||
| 1 | Ubiquinone and other terpenoid-quinone biosynthesis | |||||
| Pathwhiz Pathway | [+] 2 Pathwhiz Pathways | + | ||||
| 1 | Vitamin K Metabolism | |||||
| 2 | Coagulation | |||||
| WikiPathways | [+] 1 WikiPathways | + | ||||
| 1 | PTM: gamma carboxylation, hypusine formation and arylsulfatase activation | |||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Vitamin K antagonism of coumarin intoxication in the rat. Thromb Haemost. 1986 Apr 30;55(2):235-9. | |||||
| REF 2 | Drug information of Acenocoumarol, 2008. eduDrugs. | |||||
| REF 3 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 6808). | |||||
| REF 4 | Drug information of Dicumarol, 2008. eduDrugs. | |||||
| REF 5 | Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services. 2015 | |||||
| REF 6 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 6838). | |||||
| REF 7 | Drug information of Phenindione, 2008. eduDrugs. | |||||
| REF 8 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 6839). | |||||
| REF 9 | FDA Approved Drug Products from FDA Official Website. 2009. Application Number: (NDA) 011228. | |||||
| REF 10 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 6853). | |||||
| REF 11 | Emerging drugs in peripheral arterial disease. Expert Opin Emerg Drugs. 2006 Mar;11(1):75-90. | |||||
| REF 12 | ClinicalTrials.gov (NCT02522221) Tecarfarin Anti-Coagulation Trial (TACT). | |||||
| REF 13 | Evaluation of a reverse-hybridization StripAssay for the detection of genetic polymorphisms leading to acenocoumarol sensitivity. Mol Biol Rep. 2010 Apr;37(4):1693-7. | |||||
| REF 14 | Genotypes associated with reduced activity of VKORC1 and CYP2C9 and their modification of acenocoumarol anticoagulation during the initial treatmen... Clin Pharmacol Ther. 2009 Apr;85(4):379-86. | |||||
| REF 15 | [Oral anticoagulation and pharmacogenetics: importance in the clinical setting]. Rev Med Suisse. 2007 Sep 12;3(124):2030, 2033-4, 2036. | |||||
| REF 16 | Tecarfarin, a novel vitamin K reductase antagonist, is not affected by CYP2C9 and CYP3A4 inhibition following concomitant administration of fluconazole in healthy participants. J Clin Pharmacol. 2011Apr;51(4):561-74. | |||||
| REF 17 | Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation. Science. 2021 Jan 1;371(6524):eabc5667. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

