Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T06455
(Former ID: TTDR01443)
|
|||||
| Target Name |
Cytochrome P450 2A6 (CYP2A6)
|
|||||
| Synonyms |
Cytochrome P450(I); Cytochrome P450 IIA3; Coumarin 7-hydroxylase; CYPIIA6; CYP2A6; CYP2A3
Click to Show/Hide
|
|||||
| Gene Name |
CYP2A6
|
|||||
| Target Type |
Successful target
|
[1] | ||||
| Disease | [+] 2 Target-related Diseases | + | ||||
| 1 | Mycosis fungoides [ICD-11: 2B01] | |||||
| 2 | Psoriasis [ICD-11: EA90] | |||||
| Function |
Exhibits a high coumarin 7-hydroxylase activity. Can act in the hydroxylation of the anti-cancer drugs cyclophosphamide and ifosphamide. Competent in the metabolic activation of aflatoxin B1. Constitutes the major nicotine C-oxidase. Possesses low phenacetin O-deethylation activity.
Click to Show/Hide
|
|||||
| BioChemical Class |
Paired donor oxygen oxidoreductase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 1.14.13.-
|
|||||
| Sequence |
MLASGMLLVALLVCLTVMVLMSVWQQRKSKGKLPPGPTPLPFIGNYLQLNTEQMYNSLMK
ISERYGPVFTIHLGPRRVVVLCGHDAVREALVDQAEEFSGRGEQATFDWVFKGYGVVFSN GERAKQLRRFSIATLRDFGVGKRGIEERIQEEAGFLIDALRGTGGANIDPTFFLSRTVSN VISSIVFGDRFDYKDKEFLSLLRMMLGIFQFTSTSTGQLYEMFSSVMKHLPGPQQQAFQL LQGLEDFIAKKVEHNQRTLDPNSPRDFIDSFLIRMQEEEKNPNTEFYLKNLVMTTLNLFI GGTETVSTTLRYGFLLLMKHPEVEAKVHEEIDRVIGKNRQPKFEDRAKMPYMEAVIHEIQ RFGDVIPMSLARRVKKDTKFRDFFLPKGTEVYPMLGSVLRDPSFFSNPQDFNPQHFLNEK GQFKKSDAFVPFSIGKRNCFGEGLARMELFLFFTTVMQNFRLKSSQSPKDIDVSPKHVGF ATIPRNYTMSFLPR Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| ADReCS ID | BADD_A02053 | |||||
| HIT2.0 ID | T12BGL | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Approved Drug(s) | [+] 1 Approved Drugs | + | ||||
| 1 | Methoxsalen | Drug Info | Approved | Cutaneous T-cell lymphoma | [2], [3] | |
| Mode of Action | [+] 1 Modes of Action | + | ||||
| Binder | [+] 1 Binder drugs | + | ||||
| 1 | Methoxsalen | Drug Info | [4] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Methoxsalen | Ligand Info | |||||
| Structure Description | Crystal Structure of Human Microsomal P450 2A6 with Methoxsalen Bound | PDB:1Z11 | ||||
| Method | X-ray diffraction | Resolution | 2.05 Å | Mutation | No | [5] |
| PDB Sequence |
KGKLPPGPTP
39 LPFIGNYLQL49 NTEQMYNSLM59 KISERYGPVF69 TIHLGPRRVV79 VLCGHDAVRE 89 ALVDQAEEFS99 GRGEQATFDW109 VFKGYGVVFS119 NGERAKQLRR129 FSIATLRDFG 139 VGKRGIEERI149 QEEAGFLIDA159 LRGTGGANID169 PTFFLSRTVS179 NVISSIVFGD 189 RFDYKDKEFL199 SLLRMMLGIF209 QFTSTSTGQL219 YEMFSSVMKH229 LPGPQQQAFQ 239 LLQGLEDFIA249 KKVEHNQRTL259 DPNSPRDFID269 SFLIRMQEEE279 KNPNTEFYLK 289 NLVMTTLNLF299 IGGTETVSTT309 LRYGFLLLMK319 HPEVEAKVHE329 EIDRVIGKNR 339 QPKFEDRAKM349 PYMEAVIHEI359 QRFGDVIPMS369 LARRVKKDTK379 FRDFFLPKGT 389 EVYPMLGSVL399 RDPSFFSNPQ409 DFNPQHFLNE419 KGQFKKSDAF429 VPFSIGKRNC 439 FGEGLARMEL449 FLFFTTVMQN459 FRLKSSQSPK469 DIDVSPKHVG479 FATIPRNYTM 489 SFLPRHH
|
|||||
|
|
||||||
| Ligand Name: Pilocarpine | Ligand Info | |||||
| Structure Description | Human Cytochrome P450 2A6 I208S/I300F/G301A/S369G in complex with Pilocarpine | PDB:3T3Q | ||||
| Method | X-ray diffraction | Resolution | 2.10 Å | Mutation | Yes | [6] |
| PDB Sequence |
KLPPGPTPLP
41 FIGNYLQLNT51 EQMYNSLMKI61 SERYGPVFTI71 HLGPRRVVVL81 CGHDAVREAL 91 VDQAEEFSGR101 GEQATFDWVF111 KGYGVVFSNG121 ERAKQLRRFS131 IATLRDFGVG 141 KRGIEERIQE151 EAGFLIDALR161 GTGGANIDPT171 FFLSRTVSNV181 ISSIVFGDRF 191 DYKDKEFLSL201 LRMMLGSFQF211 TSTSTGQLYE221 MFSSVMKHLP231 GPQQQAFQLL 241 QGLEDFIAKK251 VEHNQRTLDP261 NSPRDFIDSF271 LIRMQEEEKN281 PNTEFYLKNL 291 VMTTLNLFFA301 GTETVSTTLR311 YGFLLLMKHP321 EVEAKVHEEI331 DRVIGKNRQP 341 KFEDRAKMPY351 MEAVIHEIQR361 FGDVIPMGLA371 RRVKKDTKFR381 DFFLPKGTEV 391 YPMLGSVLRD401 PSFFSNPQDF411 NPQHFLNEKG421 QFKKSDAFVP431 FSIGKRNCFG 441 EGLARMELFL451 FFTTVMQNFR461 LKSSQSPKDI471 DVSPKHVGFA481 TIPRNYTMSF 491 LPR
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Caffeine metabolism | hsa00232 | Affiliated Target |
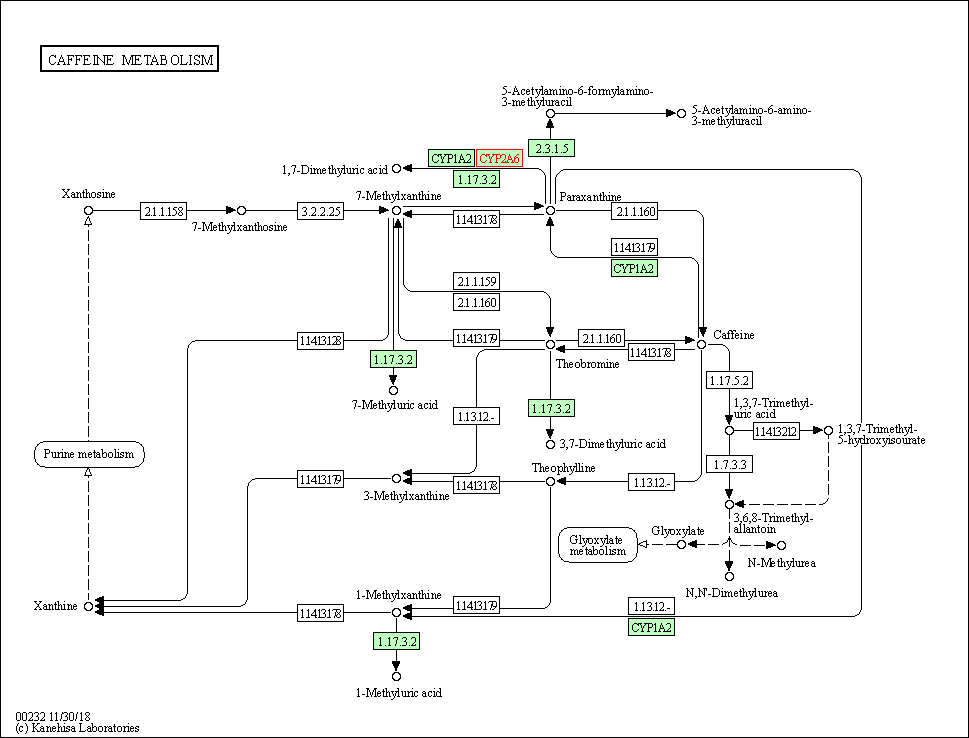
|
| Class: Metabolism => Biosynthesis of other secondary metabolites | Pathway Hierarchy | ||
| Retinol metabolism | hsa00830 | Affiliated Target |
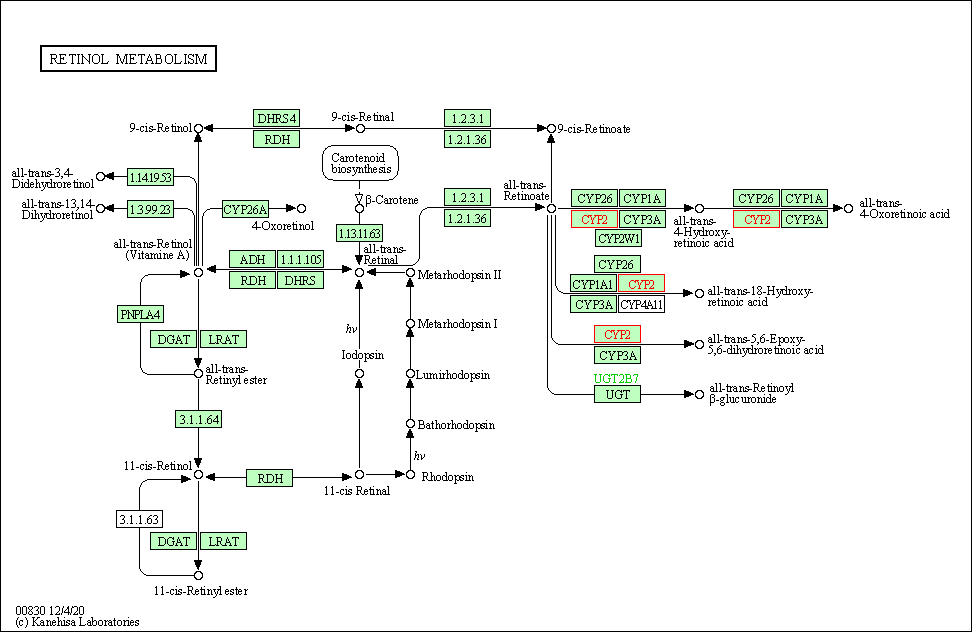
|
| Class: Metabolism => Metabolism of cofactors and vitamins | Pathway Hierarchy | ||
| Metabolism of xenobiotics by cytochrome P450 | hsa00980 | Affiliated Target |
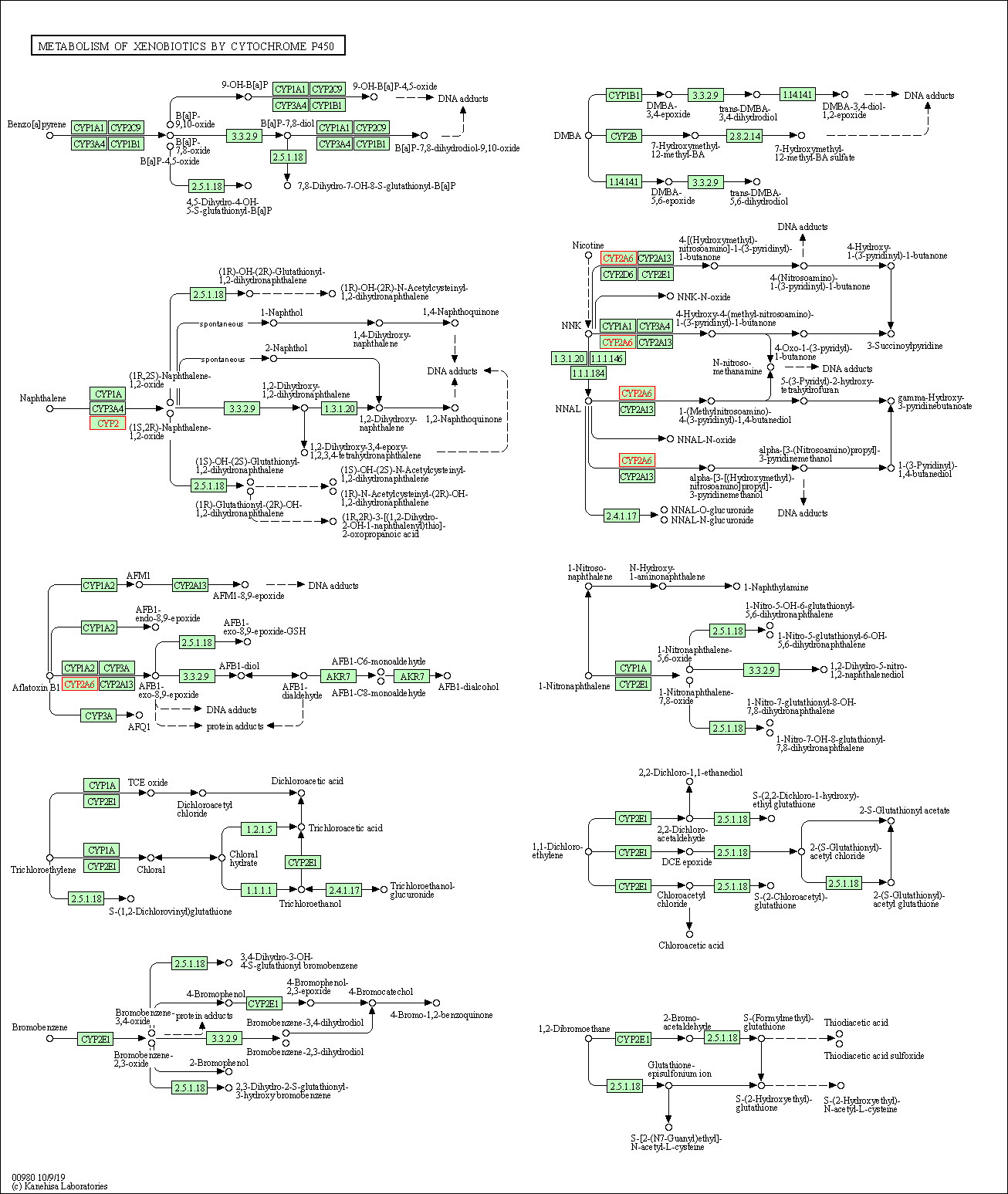
|
| Class: Metabolism => Xenobiotics biodegradation and metabolism | Pathway Hierarchy | ||
| Drug metabolism - cytochrome P450 | hsa00982 | Affiliated Target |
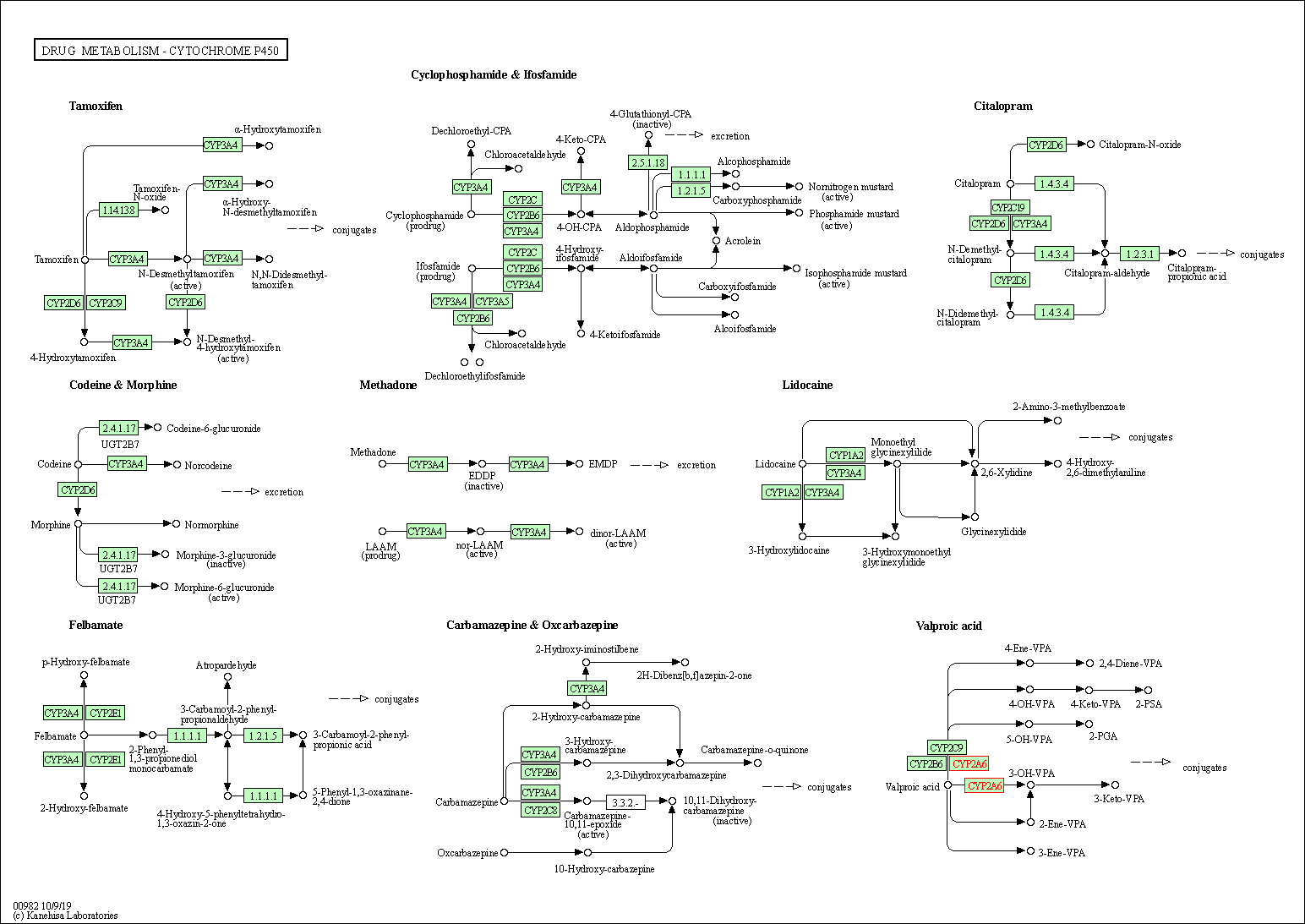
|
| Class: Metabolism => Xenobiotics biodegradation and metabolism | Pathway Hierarchy | ||
| Drug metabolism - other enzymes | hsa00983 | Affiliated Target |
|
| Class: Metabolism => Xenobiotics biodegradation and metabolism | Pathway Hierarchy | ||
| Degree | 7 | Degree centrality | 7.52E-04 | Betweenness centrality | 1.22E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 1.72E-01 | Radiality | 1.28E+01 | Clustering coefficient | 4.76E-02 |
| Neighborhood connectivity | 6.86E+00 | Topological coefficient | 2.36E-01 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target QSAR Model | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008 Jan;36(Database issue):D901-6. | |||||
| REF 2 | FDA Approved Drug Products from FDA Official Website. 2009. Application Number: (NDA) 009048. | |||||
| REF 3 | Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services. 2015 | |||||
| REF 4 | Mutagenicity and carcinogenicity of methoxsalen plus UV-A. Arch Dermatol. 1984 May;120(5):662-9. | |||||
| REF 5 | Structures of human microsomal cytochrome P450 2A6 complexed with coumarin and methoxsalen. Nat Struct Mol Biol. 2005 Sep;12(9):822-3. | |||||
| REF 6 | Structural comparison of cytochromes P450 2A6, 2A13, and 2E1 with pilocarpine. FEBS J. 2012 May;279(9):1621-31. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

