Drug Information
| Drug General Information | Top | |||
|---|---|---|---|---|
| Drug ID |
D01AZG
|
|||
| Former ID |
DAP000003
|
|||
| Drug Name |
Risperidone
|
|||
| Synonyms |
Belivon; Risperdal; Risperdone; Risperidal; Risperidona; Risperidonum; Risperin; Rispolept; Rispolin; Sequinan; Risperdal Consta; Risperidona [Spanish]; Risperidonum [Latin]; R 62 766; R 64766; R64766; Consta, Risperdal; KS-1106; R 64,766; R-118; R-64766; R64,766; Risperdal (TN); Risperdal M-Tab; Risperidal M-Tab; Risperidone (RIS); Risperidone, placebo; R 64 766, Risperdal, Risperidone; R-64,766; R-64-766; Risperidone [USAN:BAN:INN]; Risperidone (JAN/USAN/INN); 3-(2-(4-(6-Fluoro-1,2-benzisoxazol-3-yl)piperidino)ethyl)-6,7,8,9-tetrahydro-2-methyl-4H-pyrido(1,2-a)pyrimidin-4-one; 3-[2-[-4-(6-fluoro-1,2-benzisoxazol-3-yl)-1-piperidinyl]ethyl]-6,7,8,9-tetrahydro-2-methyl-4H-pyrido[1,2-a]pyrimidin-4-one; 3-[2-[4-(6-Fluoro-1,2-benzisoxazol-3-yl)-1-piperidinyl]-6,7,8,9-tetrahydro-2-methyl-4H-pyrido [1,2-a] pyrimidin-4-one; 3-[2-[4-(6-Fluoro-1,2-benzisoxazol-3-yl)piperi-dino]ethyl]-6,7,8,9-tetrahydro-2-methyl-4H-pyrido[1,2-a]-pyrimidin-4-one; 3-[2-[4-(6-fluoro-1,2-benzisoxazol-3-yl)-1-piperidinyl]ethyl]-6,7,8,9-tetrahydro-2-methyl-4H-pyrido[1,2-a]pyrimidin-4-one; 3-[2-[4-(6-fluoro-1,2-benzoxazol-3-yl)piperidin-1-yl]ethyl]-2-methyl-6,7,8,9-tetrahydropyrido[1,2-a]pyrimidin-4-one; 3-{2-[4-(6-Fluoro-benzo[d]isoxazol-3-yl)-piperidin-1-yl]-ethyl}-2-methyl-6,7,8,9-tetrahydropyrido[1,2-a]pyrimidin-4-one; 3-{2-[4-(6-fluoro-1,2-benzisoxazol-3-yl)piperidin-1-yl]ethyl}-2-methyl-6,7,8,9-tetrahydro-4H-pyrido[1,2-a]pyrimidin-4-one; 3-{2-[4-(6-fluoro-1,2-benzoxazol-3-yl)piperidin-1-yl]ethyl}-2-methyl-6,7,8,9-tetrahydro-4H-pyrido[1,2-a]pyrimidin-4-one
Click to Show/Hide
|
|||
| Drug Type |
Small molecular drug
|
|||
| Indication | Schizophrenia [ICD-11: 6A20] | Approved | [1], [2] | |
| Therapeutic Class |
Antipsychotic Agents
|
|||
| Company |
Janssen-Cilag
|
|||
| Structure |
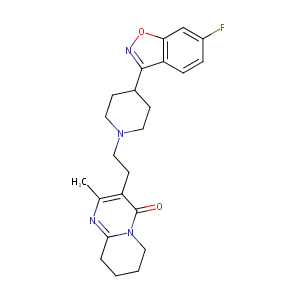 |
Download2D MOL |
||
| Formula |
C23H27FN4O2
|
|||
| Canonical SMILES |
CC1=C(C(=O)N2CCCCC2=N1)CCN3CCC(CC3)C4=NOC5=C4C=CC(=C5)F
|
|||
| InChI |
1S/C23H27FN4O2/c1-15-18(23(29)28-10-3-2-4-21(28)25-15)9-13-27-11-7-16(8-12-27)22-19-6-5-17(24)14-20(19)30-26-22/h5-6,14,16H,2-4,7-13H2,1H3
|
|||
| InChIKey |
RAPZEAPATHNIPO-UHFFFAOYSA-N
|
|||
| PubChem Compound ID | ||||
| PubChem Substance ID |
631811, 7379882, 7847492, 7980523, 8153122, 11111724, 11111725, 11467057, 11468177, 11486814, 11528727, 12013967, 14904216, 17405630, 25819945, 26719830, 26753640, 29224142, 46386613, 46505850, 49658633, 49666400, 49681677, 49699257, 49830253, 50106974, 50106975, 50123965, 53778180, 53790689, 57322595, 80652519, 81092846, 85089206, 85170980, 85209258, 85231208, 85788353, 89736125, 92126063, 92303273, 92308377, 92308738, 92712792, 93166420, 93617025, 103170825, 103856459, 103914673, 103916252
|
|||
| ChEBI ID |
CHEBI:8871
|
|||
| ADReCS Drug ID | BADD_D01951 | |||
| SuperDrug ATC ID |
N05AX08
|
|||
| SuperDrug CAS ID |
cas=106266062
|
|||
| Interaction between the Drug and Microbe | Top | |||
|---|---|---|---|---|
| The Metabolism of Drug Affected by Studied Microbe(s) | ||||
| The Order in the Taxonomic Hierarchy of the following Microbe(s): Bacteroidales | ||||
|
Studied Microbe: Alistipes indistinctus DSM22520
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Risperidone can be metabolized by Alistipes indistinctus DSM22520 (log2FC = -1.134; p = 0.006). | |||
|
Studied Microbe: Bacteroides caccae ATCC 43185
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Risperidone can be metabolized by Bacteroides caccae ATCC 43185 (log2FC = -1.03; p = 0.006). | |||
|
Studied Microbe: Bacteroides fragilis ATCC43859
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Risperidone can be metabolized by Bacteroides fragilis ATCC43859 (log2FC = -1.674; p = 0.029). | |||
|
Studied Microbe: Bacteroides fragilis HMW 610
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Risperidone can be metabolized by Bacteroides fragilis HMW 610 (log2FC = -1.015; p = 0.008). | |||
|
Studied Microbe: Bacteroides fragilis str. 3986 T(B)9
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Risperidone can be metabolized by Bacteroides fragilis str. 3986 T(B)9 (log2FC = -0.756; p = 0.005). | |||
|
Studied Microbe: Bacteroides fragilis str. DS-208
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Risperidone can be metabolized by Bacteroides fragilis str. DS-208 (log2FC = -1.091; p = 0.007). | |||
|
Studied Microbe: Odoribacter splanchnicus
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Risperidone can be metabolized by Odoribacter splanchnicus (log2FC = -0.897; p = 0.005). | |||
| The Order in the Taxonomic Hierarchy of the following Microbe(s): Bifidobacteriales | ||||
|
Studied Microbe: Bifidobacterium breve DSM 20213
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Risperidone can be metabolized by Bifidobacterium breve DSM 20213 (log2FC = -1.073; p = 0.002). | |||
| The Order in the Taxonomic Hierarchy of the following Microbe(s): Coriobacteriales | ||||
|
Studied Microbe: Collinsella intestinalis DSM 13280
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Risperidone can be metabolized by Collinsella intestinalis DSM 13280 (log2FC = -0.801; p = 0.009). | |||
| The Order in the Taxonomic Hierarchy of the following Microbe(s): Enterobacterales | ||||
|
Studied Microbe: Enterobacter cancerogenus ATCC35316
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Risperidone can be metabolized by Enterobacter cancerogenus ATCC35316 (log2FC = -0.598; p = 0.026). | |||
| The Order in the Taxonomic Hierarchy of the following Microbe(s): Erysipelotrichales | ||||
|
Studied Microbe: Eubacterium biforme DSM 3989
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Risperidone can be metabolized by Eubacterium biforme DSM 3989 (log2FC = -0.974; p = 0.005). | |||
| The Order in the Taxonomic Hierarchy of the following Microbe(s): Eubacteriales | ||||
|
Studied Microbe: Anaerotruncus colihominis DSM 17241
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Risperidone can be metabolized by Anaerotruncus colihominis DSM 17241 (log2FC = -1.505; p = 0.0). | |||
|
Studied Microbe: Blautia hansenii DSM20583
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Risperidone can be metabolized by Blautia hansenii DSM20583 (log2FC = -1.342; p = 0.001). | |||
|
Studied Microbe: Blautia luti DSM 14534
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Risperidone can be metabolized by Blautia luti DSM 14534 (log2FC = -2.054; p = 0.005). | |||
|
Studied Microbe: Bryantia formatexigens DSM 14469
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Risperidone can be metabolized by Bryantia formatexigens DSM 14469 (log2FC = -0.863; p = 0.033). | |||
|
Studied Microbe: Clostridium asparagiforme DSM 15981
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Risperidone can be metabolized by Clostridium asparagiforme DSM 15981 (log2FC = -2.178; p = 0.005). | |||
|
Studied Microbe: Clostridium difficile 120
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Risperidone can be metabolized by Clostridium difficile 120 (log2FC = -1.19; p = 0.023). | |||
|
Studied Microbe: Clostridium hathewayi DSM 13479
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Risperidone can be metabolized by Clostridium hathewayi DSM 13479 (log2FC = -0.923; p = 0.007). | |||
|
Studied Microbe: Clostridium scindens ATCC 35704
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Risperidone can be metabolized by Clostridium scindens ATCC 35704 (log2FC = -0.962; p = 0.007). | |||
|
Studied Microbe: Clostridium sp.
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Risperidone can be metabolized by Clostridium sp. (log2FC = -1.806; p = 0.0). | |||
|
Studied Microbe: Clostridium sporogenes ATCC 15579
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Risperidone can be metabolized by Clostridium sporogenes ATCC 15579 (log2FC = -6.476; p = 0.011). | |||
|
Studied Microbe: Clostridium symbiosum ATCC 14940
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Risperidone can be metabolized by Clostridium symbiosum ATCC 14940 (log2FC = -0.892; p = 0.002). | |||
|
Studied Microbe: Coprococcus comes ATCC 27758
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Risperidone can be metabolized by Coprococcus comes ATCC 27758 (log2FC = -3.515; p = 0.001). | |||
|
Studied Microbe: Enterocloster bolteae ATCC BAA-613
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Risperidone can be metabolized by Enterocloster bolteae ATCC BAA-613 (log2FC = -0.381; p = 0.029). | |||
|
Studied Microbe: Eubacterium hallii DSM3353
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Risperidone can be metabolized by Eubacterium hallii DSM3353 (log2FC = -3.735; p = 0.004). | |||
|
Studied Microbe: Eubacterium rectale ATCC 33656
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Risperidone can be metabolized by Eubacterium rectale ATCC 33656 (log2FC = -1.898; p = 0.003). | |||
|
Studied Microbe: Roseburia intestinalis L1-82
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Risperidone can be metabolized by Roseburia intestinalis L1-82 (log2FC = -2.949; p = 0.0). | |||
|
Studied Microbe: Ruminococcus gnavus ATCC 29149
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Risperidone can be metabolized by Ruminococcus gnavus ATCC 29149 (log2FC = -0.707; p = 0.003). | |||
|
Studied Microbe: Ruminococcus lactaris ATCC 29176
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Risperidone can be metabolized by Ruminococcus lactaris ATCC 29176 (log2FC = -0.838; p = 0.004). | |||
|
Studied Microbe: Ruminococcus torques ATCC 27756
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Risperidone can be metabolized by Ruminococcus torques ATCC 27756 (log2FC = -0.381; p = 0.022). | |||
|
Studied Microbe: Subdoligranulum variabile DSM 15176
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Risperidone can be metabolized by Subdoligranulum variabile DSM 15176 (log2FC = -1.549; p = 0.003). | |||
| The Order in the Taxonomic Hierarchy of the following Microbe(s): Lactobacillales | ||||
|
Studied Microbe: Enterococcus faecalis V583
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Risperidone can be metabolized by Enterococcus faecalis V583 (log2FC = -1.901; p = 0.001). | |||
| The Order in the Taxonomic Hierarchy of the following Microbe(s): Tissierellales | ||||
|
Studied Microbe: Anaerococcus hydrogenalis DSM 7454
Show/Hide Hierarchy
|
[3] | |||
| Hierarchy | ||||
| Experimental Method | High-throughput screening | |||
| Description | Risperidone can be metabolized by Anaerococcus hydrogenalis DSM 7454 (log2FC = -3.949; p = 0.002). | |||
| The Order in the Taxonomic Hierarchy of the following Microbe(s): Gut microbiota | ||||
| Studied Microbe: Gut microbiota unspecific | [4], [5], [6] | |||
| Metabolic Reaction | Isoxazole scission and hydroxylation | |||
| Metabolic Effect | Decrease activity; Induce side effect (symptoms of parkinson disease) | |||
| Description | Risperidone can be metabolized by gut microbiota through isoxazole scission and hydroxylation, which results in the decrease of drug's activity and the induction of the drug's side effect (symptoms of parkinson disease). | |||
| References | Top | |||
|---|---|---|---|---|
| REF 1 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 96). | |||
| REF 2 | Emerging anxiolytics. Expert Opin Emerg Drugs. 2007 Nov;12(4):541-54. | |||
| REF 3 | Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature. 2019 Jun;570(7762):462-467. | |||
| REF 4 | Gut microbiota modulates drug pharmacokinetics. Drug Metab Rev. 2018 Aug;50(3):357-368. | |||
| REF 5 | Human gut microbiota plays a role in the metabolism of drugs. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2016 Sep;160(3):317-26. | |||
| REF 6 | Personalized Mapping of Drug Metabolism by the Human Gut Microbiome. Cell. 2020 Jun 25;181(7):1661-1679.e22. | |||
| REF 7 | Randomized clinical comparison of perospirone and risperidone in patients with schizophrenia: Kansai Psychiatric Multicenter Study. Psychiatry Clin Neurosci. 2009 Jun;63(3):322-8. | |||
| REF 8 | Upcoming agents for the treatment of schizophrenia: mechanism of action, efficacy and tolerability. Drugs. 2008;68(16):2269-92. | |||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

