| Drug General Information |
| Drug ID |
D03ZBT
|
| Former ID |
DNAP001679
|
| Drug Name |
Crizotinib
|
| Synonyms |
Xalkori (TN); novel ALK inhibitors
|
| Drug Type |
Small molecular drug
|
| Indication |
ALK-positive advanced or metastatic NSCLC [ICD9: 162; ICD10:C33, C34]
|
Approved |
[1],
[2]
|
|---|
| Company |
Pfizer New York, NY
|
| Structure |
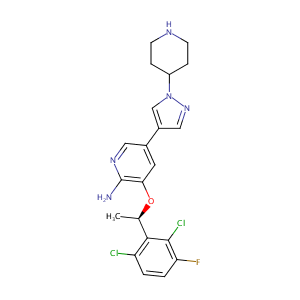
|
Download
2D MOL
3D MOL
|
| Formula |
C21H22Cl2FN5O
|
| InChI |
InChI=1S/C21H22Cl2FN5O/c1-12(19-16(22)2-3-17(24)20(19)23)30-18-8-13(9-27-21(18)25)14-10-28-29(11-14)15-4-6-26-7-5-15/h2-3,8-12,15,26H,4-7H2,1H3,(H2,25,27)/t12-/m1/s1
|
| InChIKey |
KTEIFNKAUNYNJU-GFCCVEGCSA-N
|
| CAS Number |
CAS 877399-52-5
|
| PubChem Compound ID |
|
| PubChem Substance ID |
16729581, 23733583, 74374126, 75543391, 93581001, 99309272, 99437209, 99445171, 103728028, 104161043, 119526687, 121278669, 124490471, 124756975, 125163780, 125299329, 126659900, 131407280, 135264650, 135668295, 135727397, 136345871, 136367829, 136920405, 137232008, 137275901, 141226597, 143499147, 152040652, 152134626, 152237527, 152258843, 152344161, 160644611, 160647693, 160815247, 160969694, 162010188, 162010189, 162011538, 162196520, 162927269, 163565316, 163590241, 164041842, 164193931, 165245564, 170497919, 174531154, 175265614
|
| SuperDrug ATC ID |
L01XE16
|
| Drug Resistance Mutation (DRM) |
| DRM |
DRM Info
|
| Target and Pathway |
| Target(s) |
Hepatocyte growth factor receptor |
Target Info |
Modulator |
[3]
|
|---|
| ROS1 |
Target Info |
Modulator |
[3]
|
| ALK tyrosine kinase receptor |
Target Info |
Modulator |
[3]
|
|
KEGG Pathway
|
Ras signaling pathway
|
|
Rap1 signaling pathway
|
|
Cytokine-cytokine receptor interaction
|
|
Endocytosis
|
|
PI3K-Akt signaling pathway
|
|
Axon guidance
|
|
Focal adhesion
|
|
Adherens junction
|
|
Bacterial invasion of epithelial cells
|
|
Epithelial cell signaling in Helicobacter pylori infection
|
|
Malaria
|
|
Pathways in cancer
|
|
Transcriptional misregulation in cancer
|
|
Proteoglycans in cancer
|
|
MicroRNAs in cancer
|
|
Renal cell carcinoma
|
|
Melanoma
|
|
Central carbon metabolism in cancerhsa05223:Non-small cell lung cancer
|
|
Pathway Interaction Database
|
Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met)
|
|
Arf6 signaling events
|
|
Signaling events mediated by TCPTP
|
|
Posttranslational regulation of adherens junction stability and dissassembly
|
|
Direct p53 effectors
|
|
Syndecan-1-mediated signaling events
|
|
Stabilization and expansion of the E-cadherin adherens junction
|
|
a6b1 and a6b4 Integrin signaling
|
|
FGF signaling pathway
|
|
Regulation of retinoblastoma protein
|
|
Reactome
|
Sema4D mediated inhibition of cell attachment and migration
|
|
WikiPathways
|
TGF beta Signaling Pathway
|
|
Signaling of Hepatocyte Growth Factor Receptor
|
|
Focal Adhesion
|
|
Extracellular vesicle-mediated signaling in recipient cells
|
|
Signaling Pathways in Glioblastoma
|
|
miR-targeted genes in squamous cell - TarBase
|
|
miR-targeted genes in muscle cell - TarBase
|
|
miR-targeted genes in lymphocytes - TarBase
|
|
miR-targeted genes in epithelium - TarBase
|
|
Semaphorin interactionsWP2431:Spinal Cord InjuryWP2848:Differentiation Pathway
|
| References |
| REF 1 | (http://www.guidetopharmacology.org/) Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 4903). |
|---|
| REF 2 | 2011 FDA drug approvals. Nat Rev Drug Discov. 2012 Feb 1;11(2):91-4. |
|---|
| REF 3 | Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services. 2015 |
|---|