Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T99932
|
|||||
| Target Name |
HUMAN tumor necrosis factor (TNF)
|
|||||
| Synonyms |
Tumour necrosis factor alpha; Tumour necrosis factor; Tumor necrosis factor ligand superfamily member 2; TNFalpha; TNFSF2; TNFA; TNF-alpha; TNF-a; TNF alpha; Cachectin
Click to Show/Hide
|
|||||
| Gene Name |
TNF
|
|||||
| Function |
It is mainly secreted by macrophages and can induce cell death of certain tumor cell lines. It is potent pyrogen causing fever by direct action or by stimulation of interleukin-1 secretion and is implicated in the induction of cachexia, Under certain conditions it can stimulate cell proliferation and induce cell differentiation. Impairs regulatory T-cells (Treg) function in individuals with rheumatoid arthritis via FOXP3 dephosphorylation. Upregulates the expression of protein phosphatase 1 (PP1), which dephosphorylates the key 'Ser-418' residue of FOXP3, thereby inactivating FOXP3 and rendering Treg cells functionally defective. Key mediator of cell death in the anticancer action of BCG-stimulated neutrophils in combination with DIABLO/SMAC mimetic in the RT4v6 bladder cancer cell line. Induces insulin resistance in adipocytes via inhibition of insulin-induced IRS1 tyrosine phosphorylation and insulin-induced glucose uptake. Induces GKAP42 protein degradation in adipocytes which is partially responsible for TNF-induced insulin resistance. Cytokine that binds to TNFRSF1A/TNFR1 and TNFRSF1B/TNFBR.
Click to Show/Hide
|
|||||
| BioChemical Class |
Cytokine: tumor necrosis factor
|
|||||
| UniProt ID | ||||||
| Sequence |
MSTESMIRDVELAEEALPKKTGGPQGSRRCLFLSLFSFLIVAGATTLFCLLHFGVIGPQR
EEFPRDLSLISPLAQAVRSSSRTPSDKPVAHVVANPQAEGQLQWLNRRANALLANGVELR DNQLVVPSEGLYLIYSQVLFKGQGCPSTHVLLTHTISRIAVSYQTKVNLLSAIKSPCQRE TPEGAEAKPWYEPIYLGGVFQLEKGDRLSAEINRPDYLDFAESGQVYFGIIAL Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: 4-N-benzyl-4-N-[2-[3-(2-piperazin-1-ylpyrimidin-5-yl)phenyl]phenyl]pyrimidine-2,4-diamine | Ligand Info | |||||
| Structure Description | HUMAN TNF-ALPHA IN COMPLEX WITH JNJ525 | PDB:5MU8 | ||||
| Method | X-ray diffraction | Resolution | 3.00 Å | Mutation | No | [2] |
| PDB Sequence |
SDKPVAHVVA
18 NPQAEGQLQW28 LNALLANGVE42 LRDNQLVVPS52 EGLYLIYSQV62 LFKGQGCPST 72 HVLLTHTISR82 IAVSYQTKVN92 LLSAIKSPCQ102 RETEAKPWYE116 PIYLGGVFQL 126 EKGDRLSAEI136 NRPDYLDFAE146 SGQVYFGIIA156 L
|
|||||
|
|
||||||
| Ligand Name: (1-Oxyl-2,2,5,5-tetramethylpyrroline-3-methyl)methanethiosulfonate | Ligand Info | |||||
| Structure Description | Crystal Structure of Spin-Labeled T77C TNFa | PDB:5UUI | ||||
| Method | X-ray diffraction | Resolution | 1.40 Å | Mutation | Yes | [3] |
| PDB Sequence |
SDKPVAHVVA
18 NPQAEGQLQW28 LNRRANALLA38 NGVELRDNQL48 VVPSEGLYLI58 YSQVLFKGQG 68 VLLCHTISRI83 AVSYQTKVNL93 LSAIKSPAKP113 WYEPIYLGGV123 FQLEKGDRLS 133 AEINRPDYLD143 FAESGQVYFG153 IIAL
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| MAPK signaling pathway | hsa04010 | Affiliated Target |
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Cytokine-cytokine receptor interaction | hsa04060 | Affiliated Target |
|
| Class: Environmental Information Processing => Signaling molecules and interaction | Pathway Hierarchy | ||
| Viral protein interaction with cytokine and cytokine receptor | hsa04061 | Affiliated Target |
|
| Class: Environmental Information Processing => Signaling molecules and interaction | Pathway Hierarchy | ||
| NF-kappa B signaling pathway | hsa04064 | Affiliated Target |
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Sphingolipid signaling pathway | hsa04071 | Affiliated Target |
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| mTOR signaling pathway | hsa04150 | Affiliated Target |
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Apoptosis | hsa04210 | Affiliated Target |
|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Necroptosis | hsa04217 | Affiliated Target |
|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| TGF-beta signaling pathway | hsa04350 | Affiliated Target |
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Osteoclast differentiation | hsa04380 | Affiliated Target |
|
| Class: Organismal Systems => Development and regeneration | Pathway Hierarchy | ||
| Antigen processing and presentation | hsa04612 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Toll-like receptor signaling pathway | hsa04620 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| NOD-like receptor signaling pathway | hsa04621 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| RIG-I-like receptor signaling pathway | hsa04622 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| C-type lectin receptor signaling pathway | hsa04625 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Hematopoietic cell lineage | hsa04640 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Natural killer cell mediated cytotoxicity | hsa04650 | Affiliated Target |
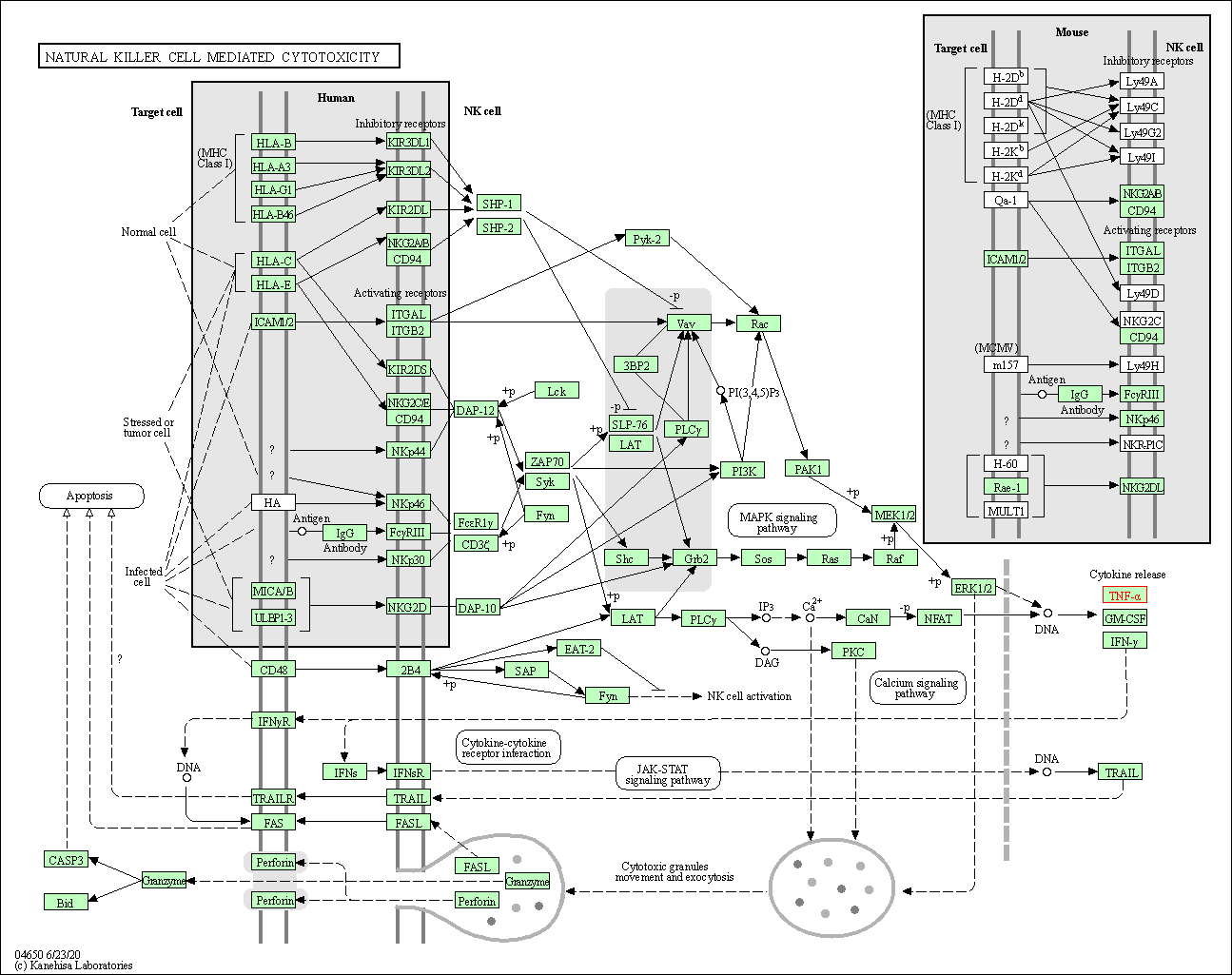
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| IL-17 signaling pathway | hsa04657 | Affiliated Target |
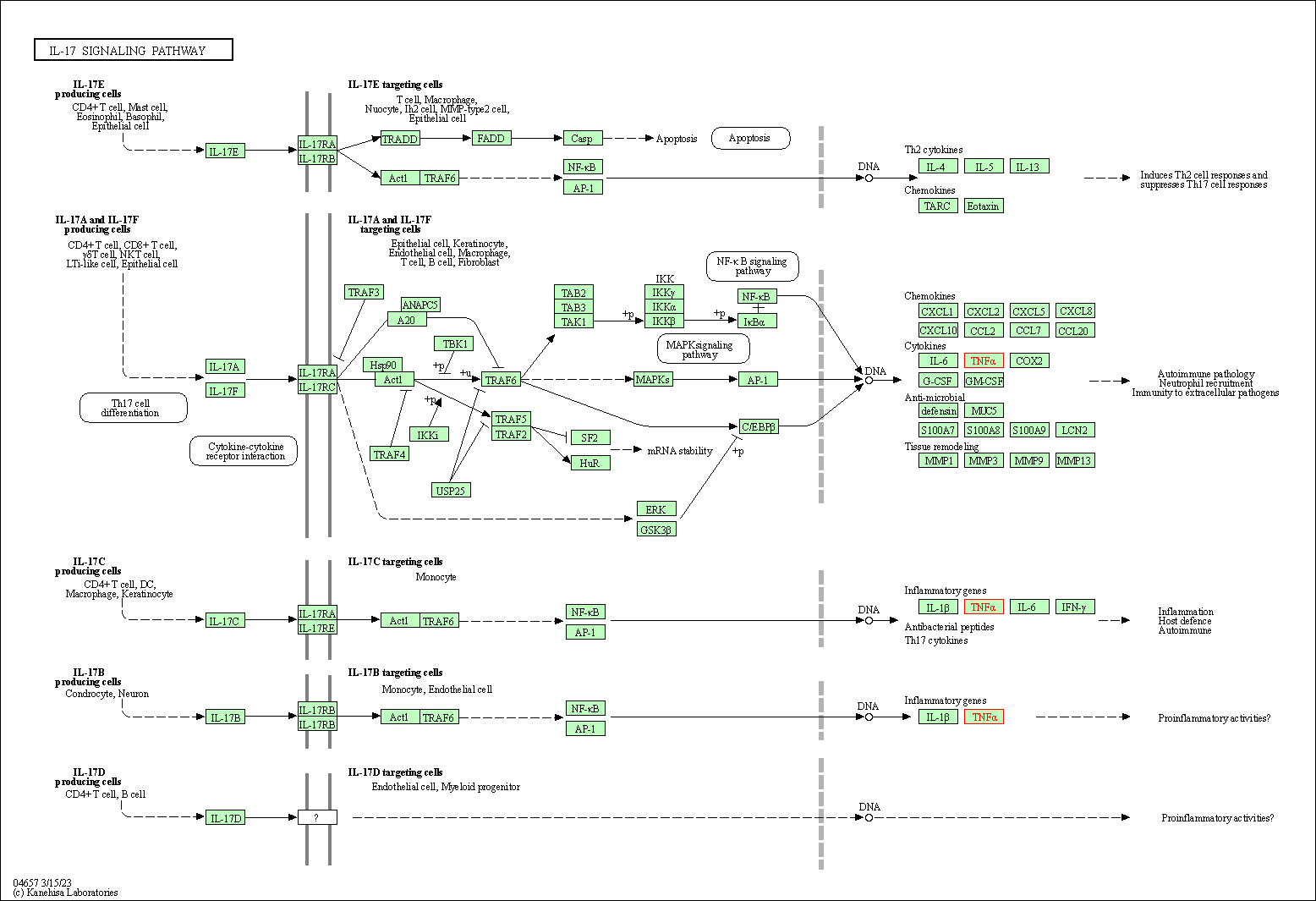
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| T cell receptor signaling pathway | hsa04660 | Affiliated Target |
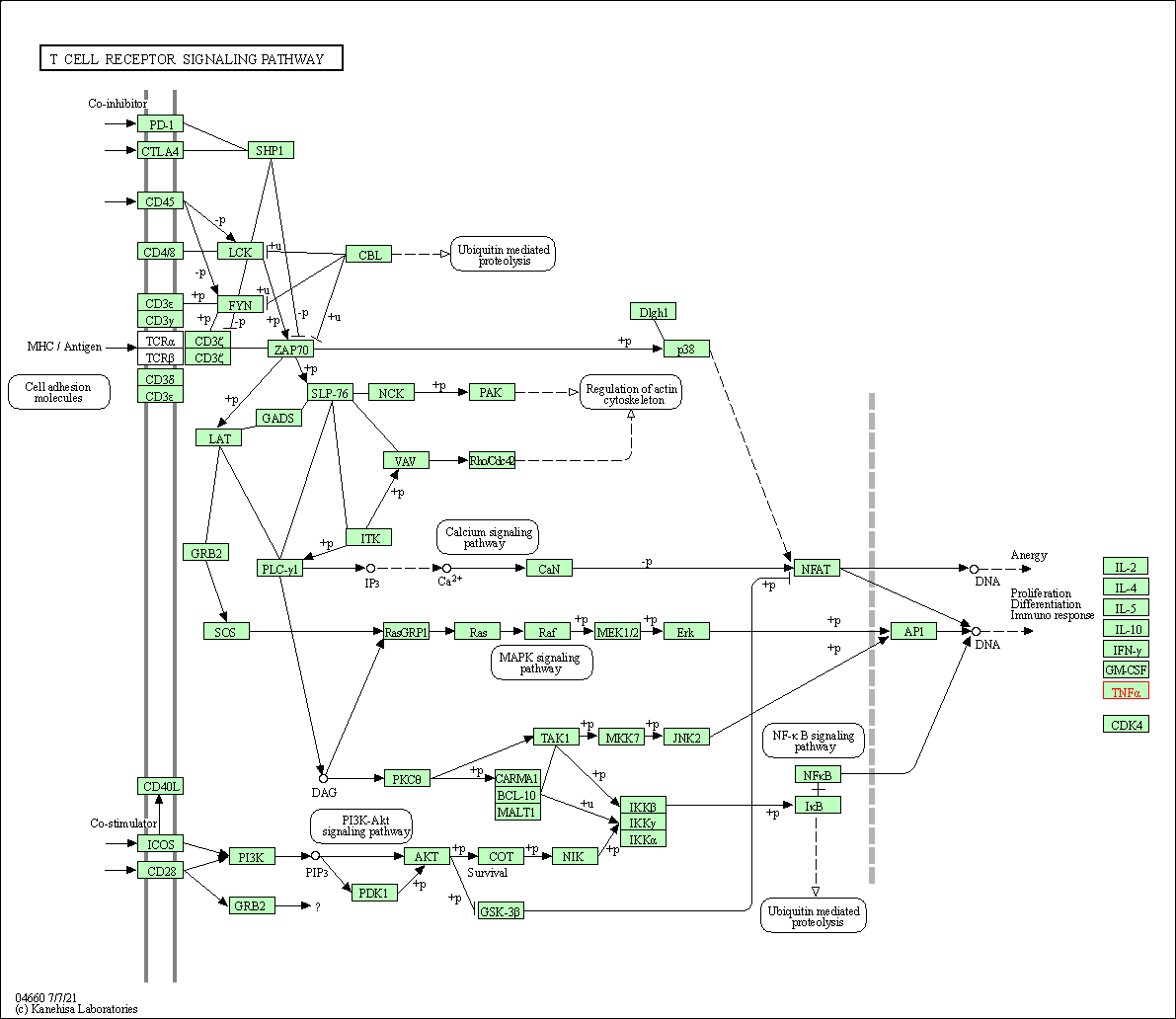
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Fc epsilon RI signaling pathway | hsa04664 | Affiliated Target |
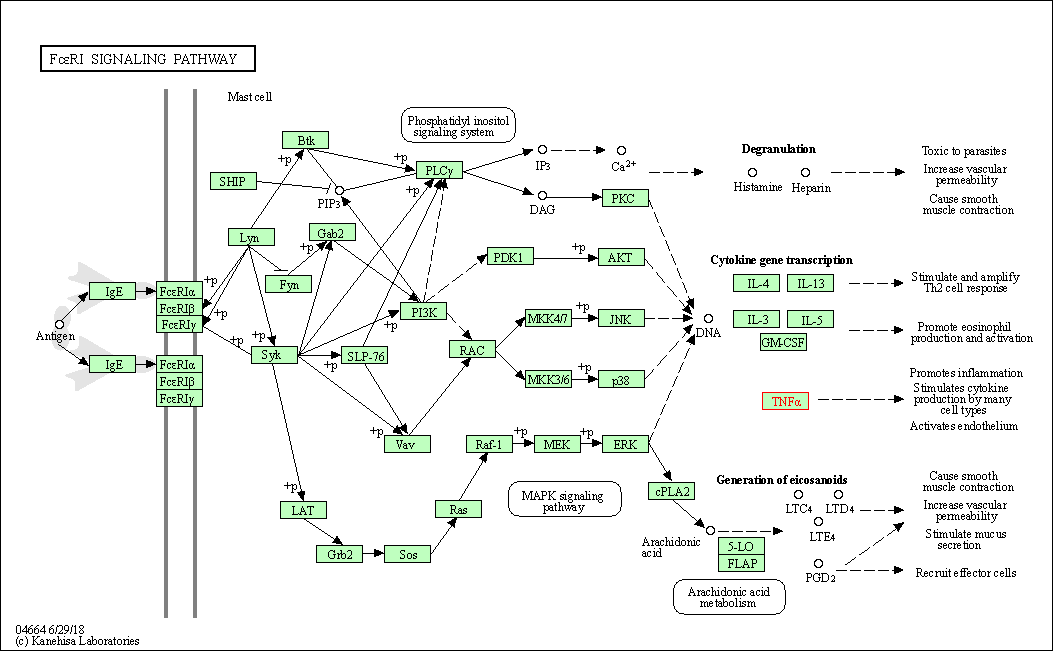
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| TNF signaling pathway | hsa04668 | Affiliated Target |
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Adipocytokine signaling pathway | hsa04920 | Affiliated Target |
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 76 | Degree centrality | 8.17E-03 | Betweenness centrality | 8.97E-03 |
|---|---|---|---|---|---|
| Closeness centrality | 2.63E-01 | Radiality | 1.45E+01 | Clustering coefficient | 1.60E-01 |
| Neighborhood connectivity | 3.63E+01 | Topological coefficient | 3.75E-02 | Eccentricity | 10 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | ClinicalTrials.gov (NCT04363008) COVID-19 Inflammatory Blood Biomarkers for Clinical Management, Prognosis and Evaluation of Interventions | |||||
| REF 2 | Structural Basis of Small-Molecule Aggregate Induced Inhibition of a Protein-Protein Interaction. J Med Chem. 2017 Apr 27;60(8):3511-3517. | |||||
| REF 3 | Natural Conformational Sampling of Human TNFAlpha Visualized by Double Electron-Electron Resonance. Biophys J. 2017 Jul 25;113(2):371-380. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

