Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T98337
(Former ID: TTDNR00722)
|
|||||
| Target Name |
FK506-binding protein 5 (FKBP5)
|
|||||
| Synonyms |
Peptidyl-prolyl cis-trans isomerase FKBP5; PPIase FKBP5; HSP90-binding immunophilin; FKBP54; FKBP51; FKBP-51; FKBP-5; FF1 antigen; Androgen-regulated protein 6; AIG6; 54 kDa progesterone receptor-associated immunophilin; 51 kDa FKBP; 51 kDa FK506-binding protein
Click to Show/Hide
|
|||||
| Gene Name |
FKBP5
|
|||||
| Target Type |
Literature-reported target
|
[1] | ||||
| Function |
Component of unligated steroid receptors heterocomplexes through interaction with heat-shock protein 90 (HSP90). Plays a role in the intracellular trafficking of heterooligomeric forms of steroid hormone receptors maintaining the complex into the cytoplasm when unliganded. Immunophilin protein with PPIase and co-chaperone activities.
Click to Show/Hide
|
|||||
| BioChemical Class |
Cis-trans-isomerase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 5.2.1.8
|
|||||
| Sequence |
MTTDEGAKNNEESPTATVAEQGEDITSKKDRGVLKIVKRVGNGEETPMIGDKVYVHYKGK
LSNGKKFDSSHDRNEPFVFSLGKGQVIKAWDIGVATMKKGEICHLLCKPEYAYGSAGSLP KIPSNATLFFEIELLDFKGEDLFEDGGIIRRTKRKGEGYSNPNEGATVEIHLEGRCGGRM FDCRDVAFTVGEGEDHDIPIGIDKALEKMQREEQCILYLGPRYGFGEAGKPKFGIEPNAE LIYEVTLKSFEKAKESWEMDTKEKLEQAAIVKEKGTVYFKGGKYMQAVIQYGKIVSWLEM EYGLSEKESKASESFLLAAFLNLAMCYLKLREYTKAVECCDKALGLDSANEKGLYRRGEA QLLMNEFESAKGDFEKVLEVNPQNKAARLQISMCQKKAKEHNERDRRIYANMFKKFAEQD AKEEANKAMGKKTSEGVTNEKGTDSQAMEEEKPEGHV Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Sirolimus | Ligand Info | |||||
| Structure Description | Co-crystal structure of the PPIase domain of FKBP51, Rapamycin and the FRB fragment of mTOR | PDB:4DRI | ||||
| Method | X-ray diffraction | Resolution | 1.45 Å | Mutation | No | [2] |
| PDB Sequence |
EQGEDITSKK
29 DRGVLKIVKR39 VGNGEETPMI49 GDKVYVHYKG59 KLSNGKKFDS69 SHDRNEPFVF 79 SLGKGQVIKA89 WDIGVATMKK99 GEICHLLCKP109 EYAYGSAGSL119 PKIPSNATLF 129 FEIELLDFKG139 E
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Ligand Name: Nicotinamide | Ligand Info | |||||
| Structure Description | CRYSTAL STRUCTURE OF HUMAN FKBP51 FK1 DOMAIN A19T MUTANT IN COMPLEX WITH NICOTINAMIDE | PDB:6TX6 | ||||
| Method | X-ray diffraction | Resolution | 0.98 Å | Mutation | Yes | [3] |
| PDB Sequence |
GAPATVTEQG
22 EDITSKKDRG32 VLKIVKRVGN42 GEETPMIGDK52 VYVHYKGKLS62 NGKKFDSSHD 72 RNEPFVFSLG82 KGQVIKAWDI92 GVATMKKGEI102 CHLLCKPEYA112 YGSAGSLPKI 122 PSNATLFFEI132 ELLDFKGE
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Estrogen signaling pathway | hsa04915 | Affiliated Target |
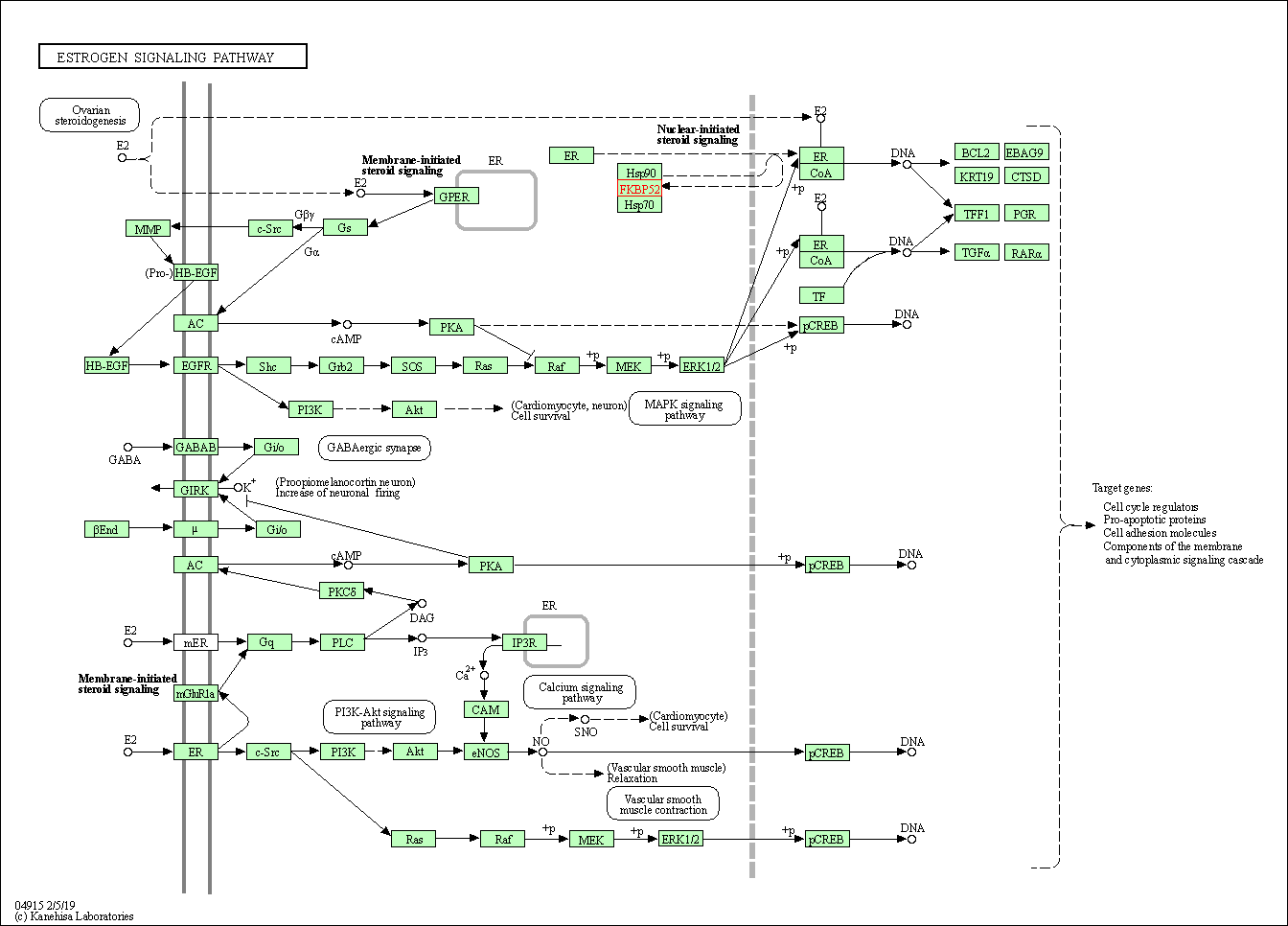
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Degree | 8 | Degree centrality | 8.59E-04 | Betweenness centrality | 1.24E-05 |
|---|---|---|---|---|---|
| Closeness centrality | 2.39E-01 | Radiality | 1.42E+01 | Clustering coefficient | 4.29E-01 |
| Neighborhood connectivity | 5.79E+01 | Topological coefficient | 1.74E-01 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-interacting Proteins | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Biomarkers for eosinophilic esophagitis: a review. Ann Allergy Asthma Immunol. 2012 Sep;109(3):155-9. | |||||
| REF 2 | Large FK506-binding proteins shape the pharmacology of rapamycin. Mol Cell Biol. 2013 Apr;33(7):1357-67. | |||||
| REF 3 | Hybrid Screening Approach for Very Small Fragments: X-ray and Computational Screening on FKBP51. J Med Chem. 2020 Jun 11;63(11):5856-5864. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

