Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T98155
|
|||||
| Target Name |
HUMAN interleukin-1 beta (IL1B)
|
|||||
| Synonyms |
IL1F2; IL-1beta; IL-1 beta; Catabolin
Click to Show/Hide
|
|||||
| Gene Name |
IL1B
|
|||||
| Function |
Initially discovered as the major endogenous pyrogen, induces prostaglandin synthesis, neutrophil influx and activation, T-cell activation and cytokine production, B-cell activation and antibody production, and fibroblast proliferation and collagen production. Promotes Th17 differentiation of T-cells. Synergizes with IL12/interleukin-12 to induce IFNG synthesis from T-helper 1 (Th1) cells. Potent proinflammatory cytokine.
Click to Show/Hide
|
|||||
| BioChemical Class |
Cytokine: interleukin
|
|||||
| UniProt ID | ||||||
| Sequence |
MAEVPELASEMMAYYSGNEDDLFFEADGPKQMKCSFQDLDLCPLDGGIQLRISDHHYSKG
FRQAASVVVAMDKLRKMLVPCPQTFQENDLSTFFPFIFEEEPIFFDTWDNEAYVHDAPVR SLNCTLRDSQQKSLVMSGPYELKALHLQGQDMEQQVVFSMSFVQGEESNDKIPVALGLKE KNLYLSCVLKDDKPTLQLESVDPKNYPKKKMEKRFVFNKIEINNKLEFESAQFPNWYIST SQAENMPVFLGGTKGGQDITDFTMQFVSS Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: N-{2-[4-(AMINOSULFONYL)PHENYL]ETHYL}ACETAMIDE | Ligand Info | |||||
| Structure Description | PanDDA analysis group deposition INTERLEUKIN-1 BETA -- Fragment Z30857828 in complex with INTERLEUKIN-1 BETA | PDB:5R8C | ||||
| Method | X-ray diffraction | Resolution | 1.54 Å | Mutation | No | [2] |
| PDB Sequence |
APVRSLNCTL
10 RDSQQKSLVM20 SGPYELKALH30 LQGQDMEQQV40 VFSMSFVQGE50 ESNDKIPVAL 60 GLKEKNLYLS70 CVLKDDKPTL80 QLESVDPKNY90 PKKKMEKRFV100 FNKIEINNKL 110 EFESAQFPNW120 YISTSQAENM130 PVFLGGTKGG140 QDITDFTMQF150 VS |
|||||
|
|
||||||
| Ligand Name: 1-methyl-N-{[(2S)-oxolan-2-yl]methyl}-1H-pyrazole-3-carboxamide | Ligand Info | |||||
| Structure Description | PanDDA analysis group deposition INTERLEUKIN-1 BETA -- Fragment Z2643472210 in complex with INTERLEUKIN-1 BETA | PDB:5R8Q | ||||
| Method | X-ray diffraction | Resolution | 1.23 Å | Mutation | No | [2] |
| PDB Sequence |
APVRSLNCTL
10 RDSQQKSLVM20 SGPYELKALH30 LQGQDMEQQV40 VFSMSFVQGE50 ESNDKIPVAL 60 GLKEKNLYLS70 CVLKDDKPTL80 QLESVDPKNY90 PKKKMEKRFV100 FNKIEINNKL 110 EFESAQFPNW120 YISTSQAENM130 PVFLGGTKGG140 QDITDFTMQF150 VS |
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| MAPK signaling pathway | hsa04010 | Affiliated Target |
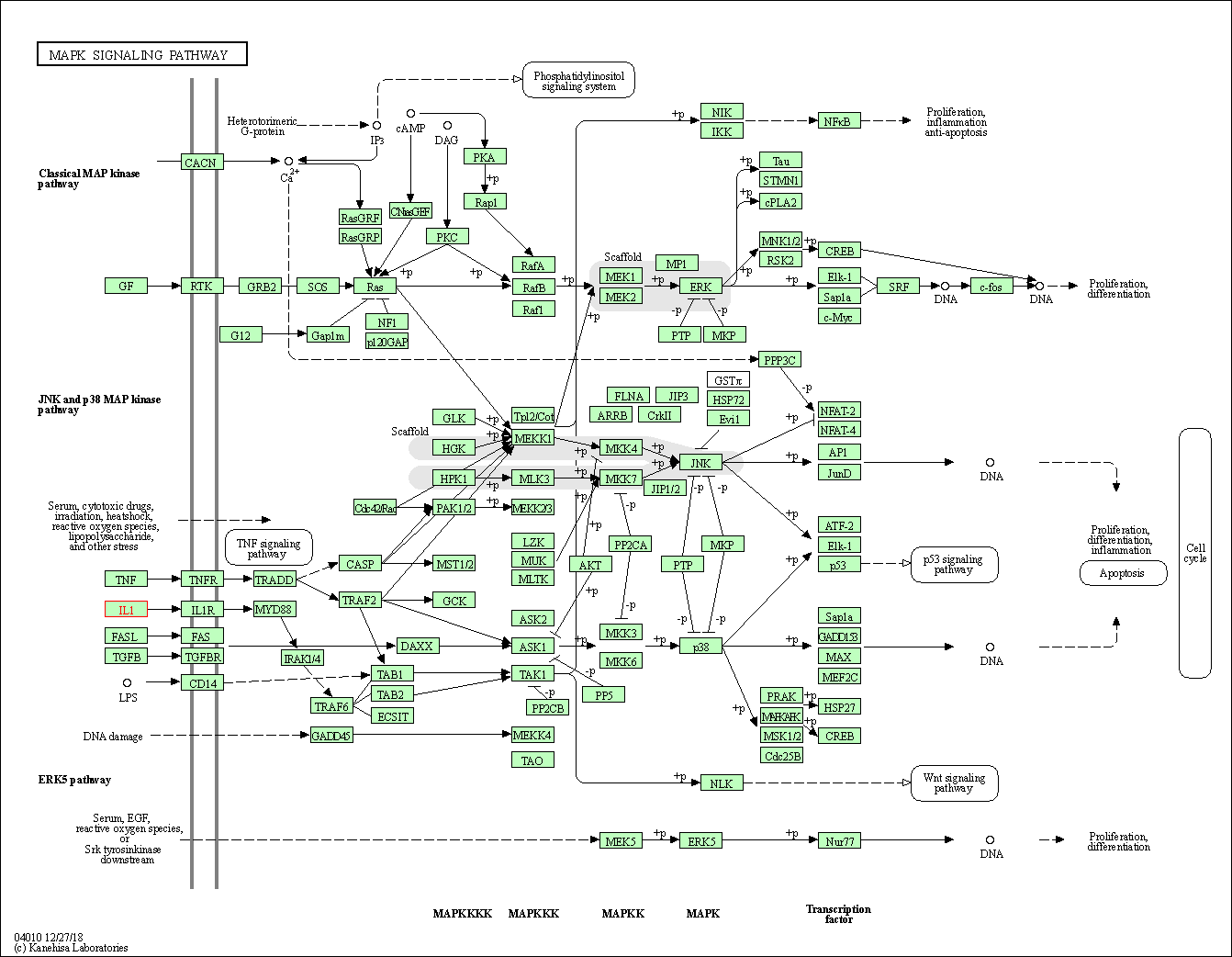
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Cytokine-cytokine receptor interaction | hsa04060 | Affiliated Target |
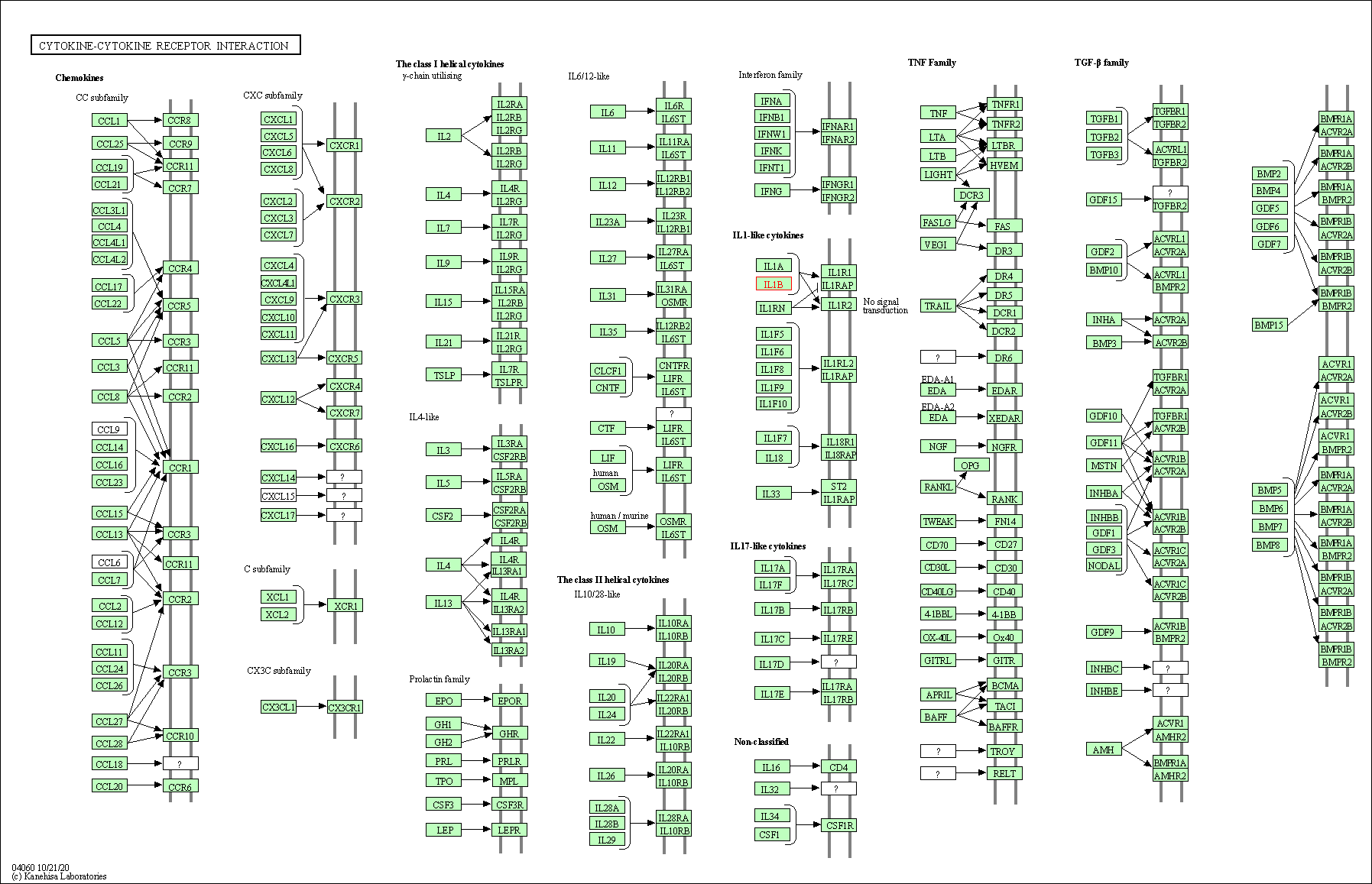
|
| Class: Environmental Information Processing => Signaling molecules and interaction | Pathway Hierarchy | ||
| NF-kappa B signaling pathway | hsa04064 | Affiliated Target |
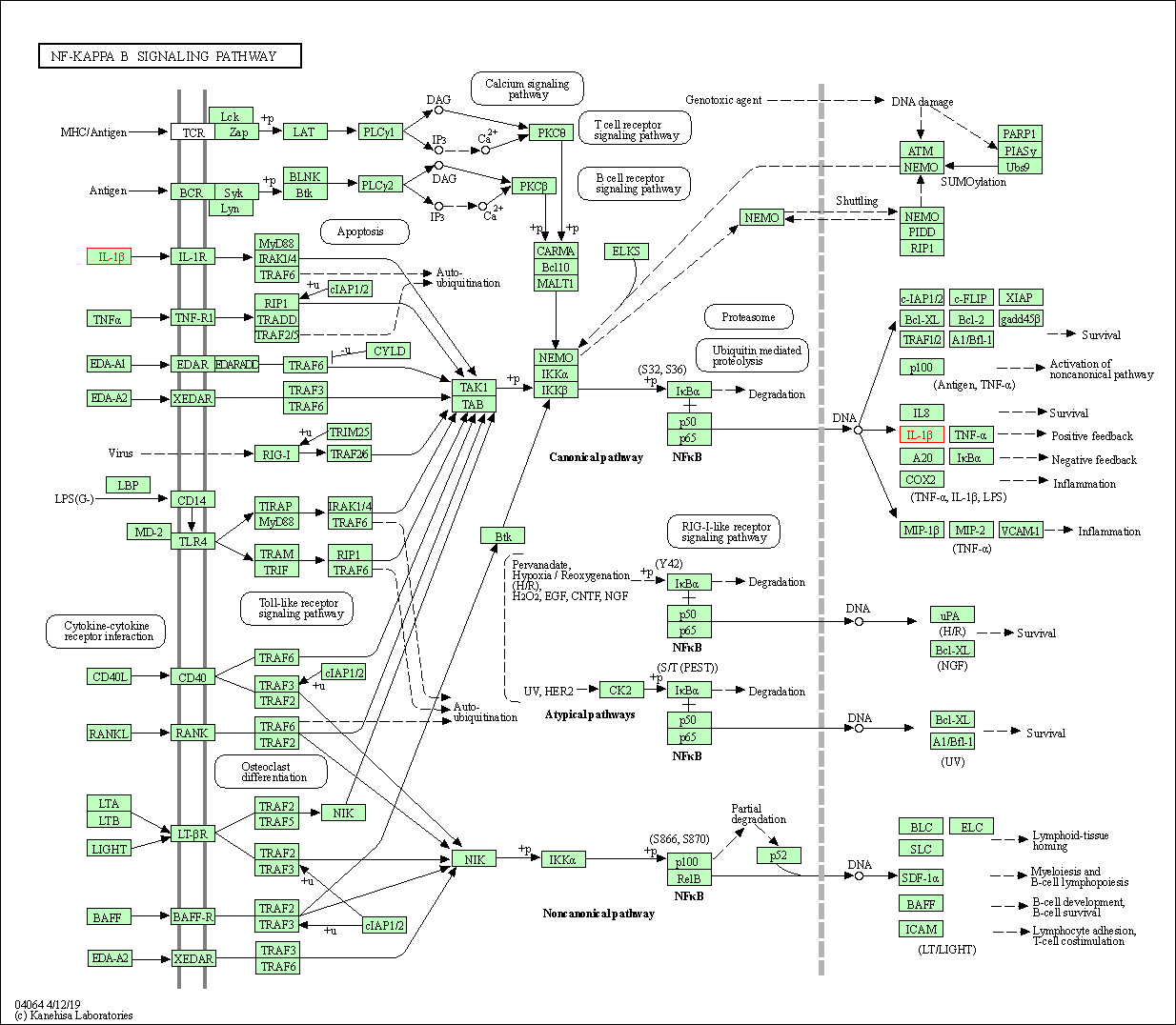
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Necroptosis | hsa04217 | Affiliated Target |
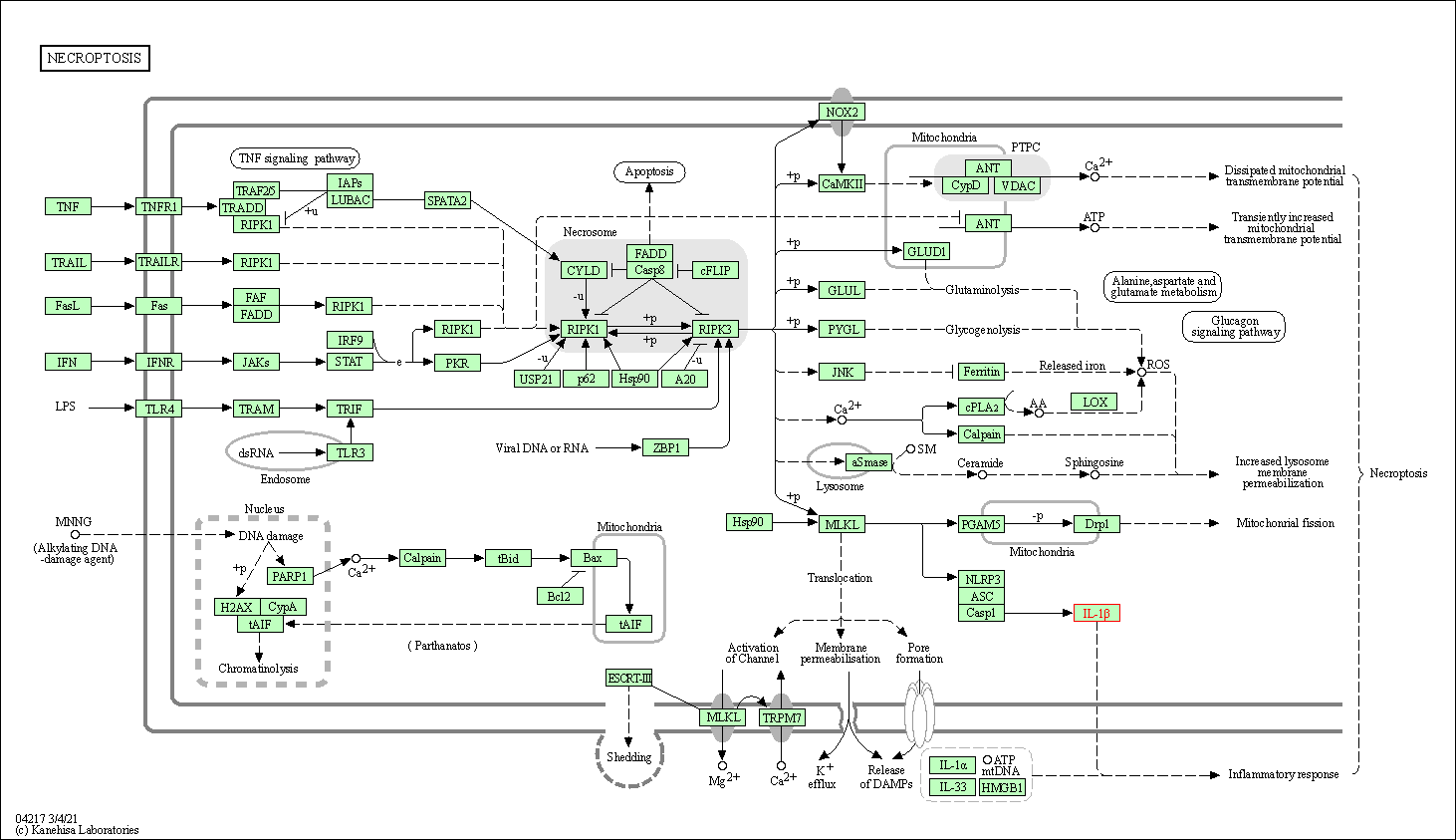
|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Osteoclast differentiation | hsa04380 | Affiliated Target |
|
| Class: Organismal Systems => Development and regeneration | Pathway Hierarchy | ||
| Toll-like receptor signaling pathway | hsa04620 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| NOD-like receptor signaling pathway | hsa04621 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Cytosolic DNA-sensing pathway | hsa04623 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| C-type lectin receptor signaling pathway | hsa04625 | Affiliated Target |
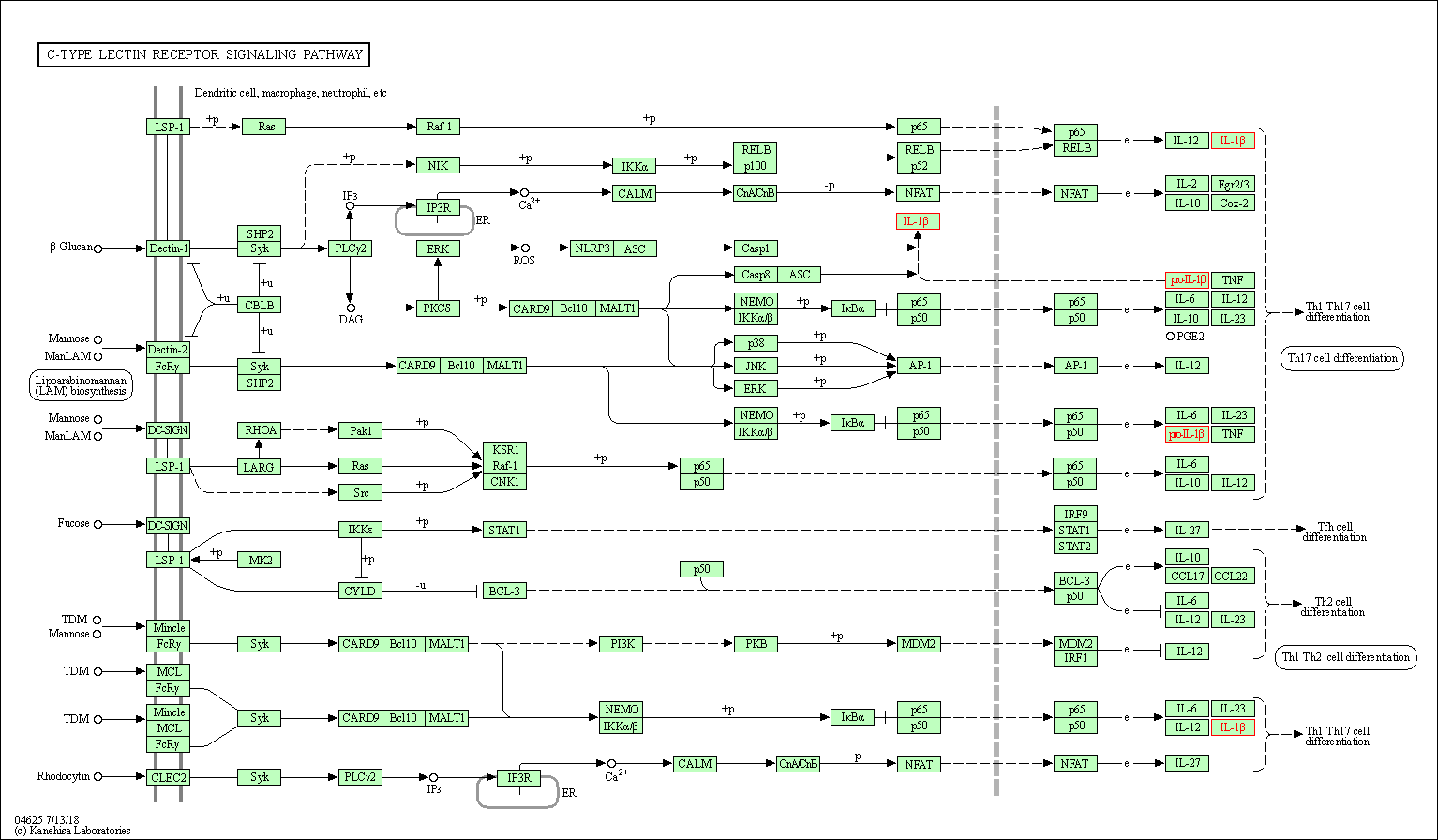
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Hematopoietic cell lineage | hsa04640 | Affiliated Target |
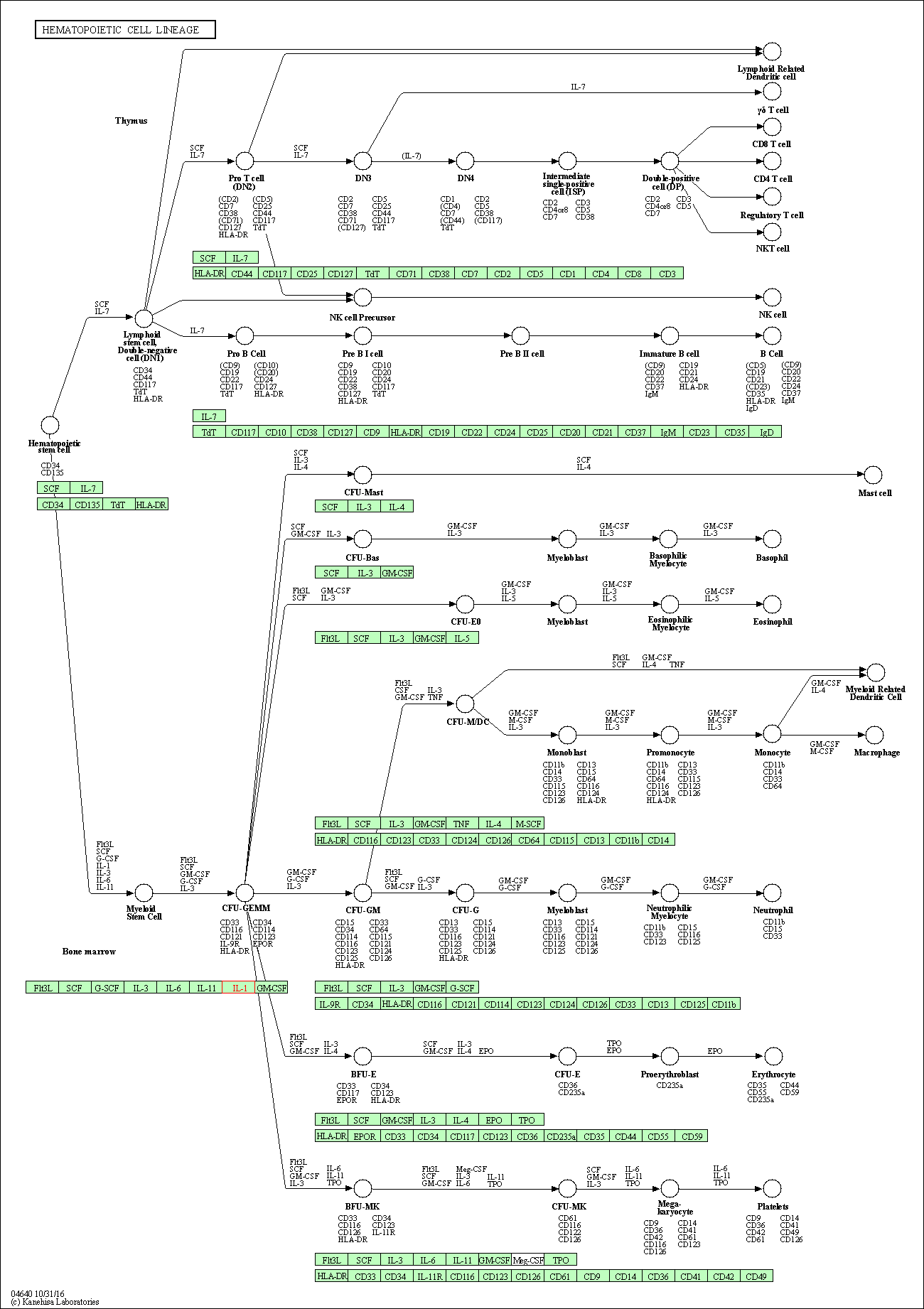
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| IL-17 signaling pathway | hsa04657 | Affiliated Target |
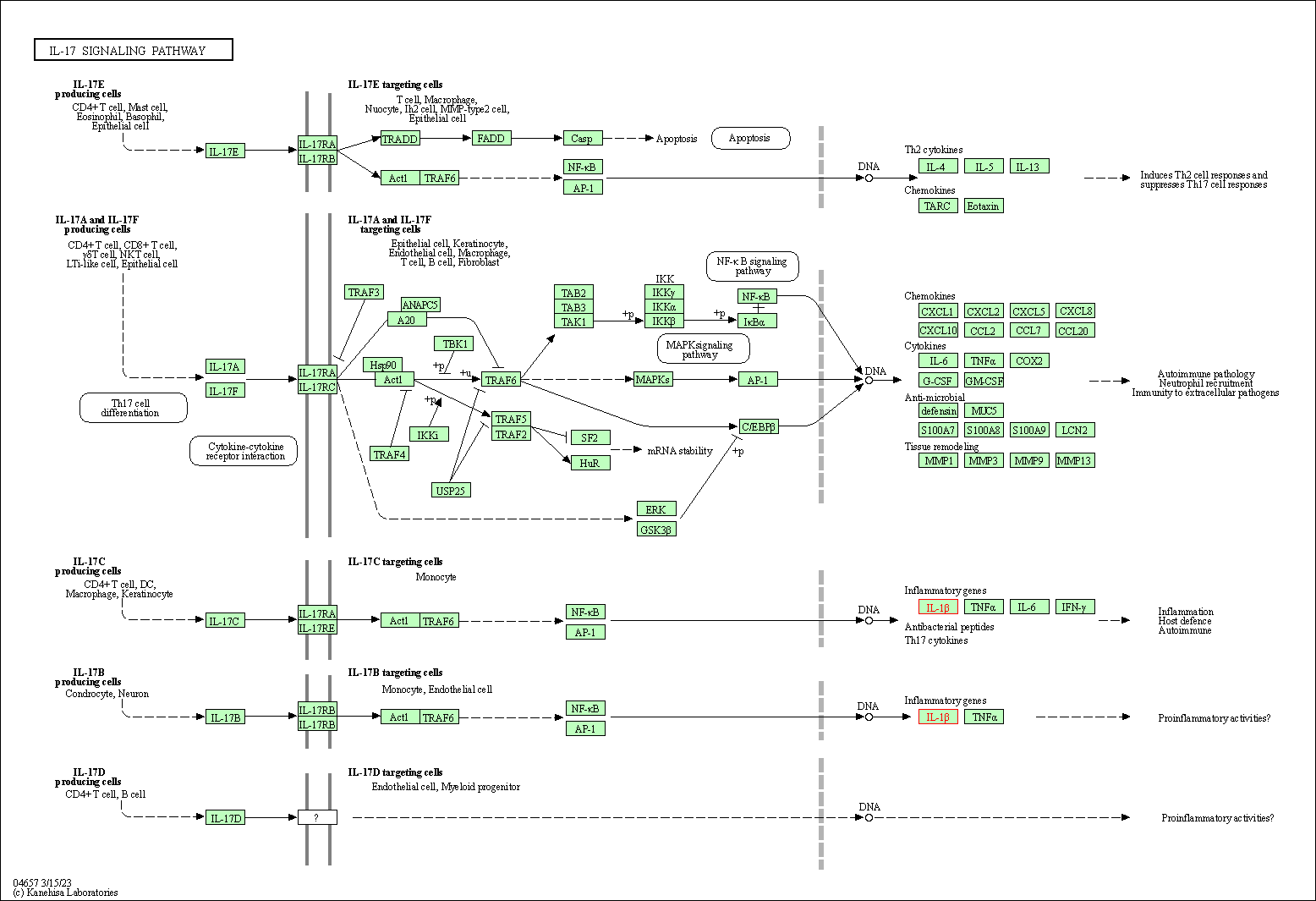
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Th17 cell differentiation | hsa04659 | Affiliated Target |
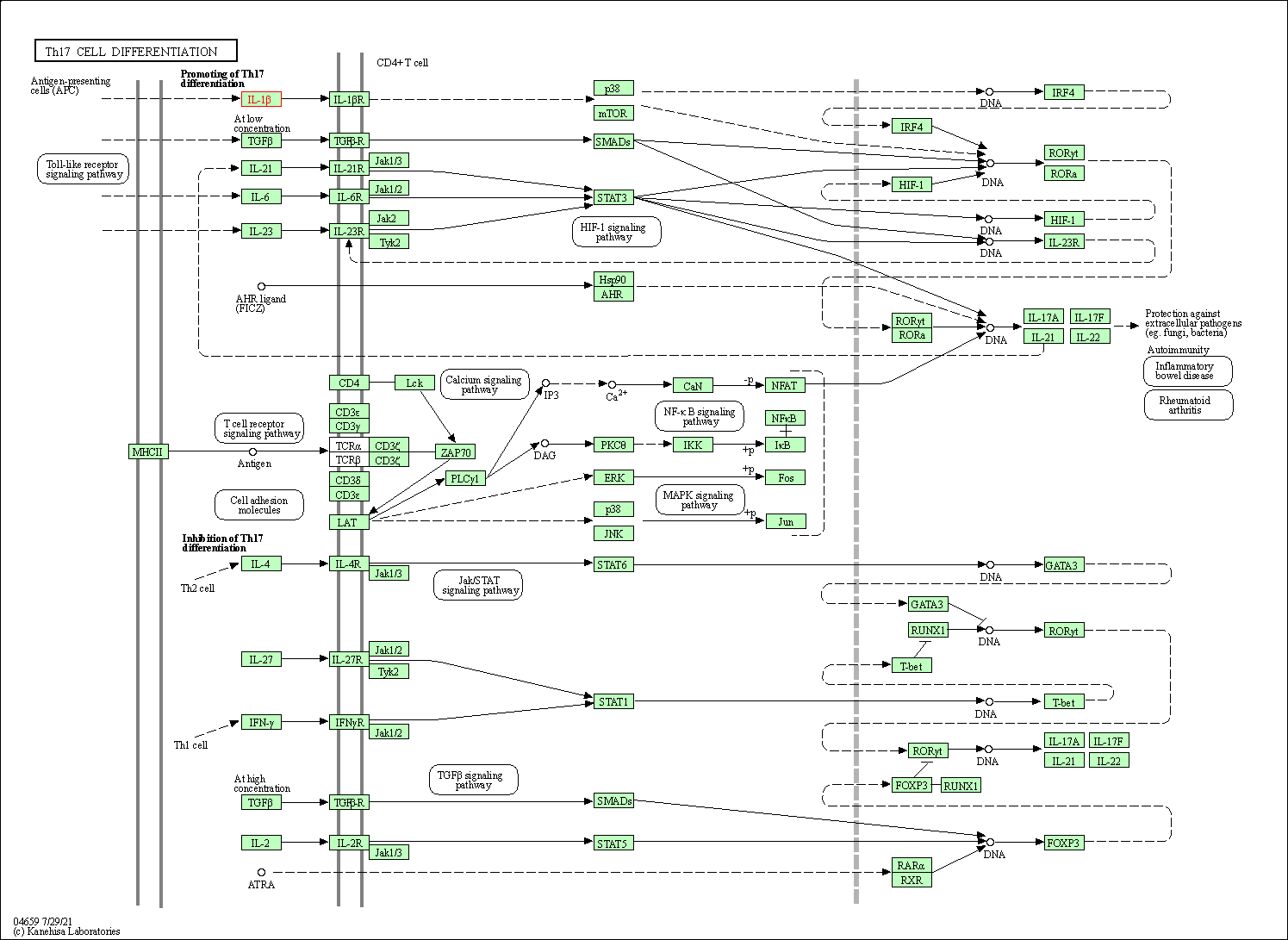
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| TNF signaling pathway | hsa04668 | Affiliated Target |
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Inflammatory mediator regulation of TRP channels | hsa04750 | Affiliated Target |
|
| Class: Organismal Systems => Sensory system | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 46 | Degree centrality | 4.94E-03 | Betweenness centrality | 1.51E-03 |
|---|---|---|---|---|---|
| Closeness centrality | 2.41E-01 | Radiality | 1.42E+01 | Clustering coefficient | 2.31E-01 |
| Neighborhood connectivity | 2.93E+01 | Topological coefficient | 5.87E-02 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | ClinicalTrials.gov (NCT04363008) COVID-19 Inflammatory Blood Biomarkers for Clinical Management, Prognosis and Evaluation of Interventions | |||||
| REF 2 | Mining the PDB for Tractable Cases Where X-ray Crystallography Combined with Fragment Screens Can Be Used to Systematically Design Protein-Protein Inhibitors: Two Test Cases Illustrated by IL1beta-IL1R and p38Alpha-TAB1 Complexes. J Med Chem. 2020 Jul 23;63(14):7559-7568. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

