Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T98022
(Former ID: TTDC00053)
|
|||||
| Target Name |
Carboxypeptidase B2 (CPB2)
|
|||||
| Synonyms |
Thrombin-activable fibrinolysis inhibitor; TAFI; Plasma carboxypeptidase B; Carboxypeptidase U; CPU; CPB2
Click to Show/Hide
|
|||||
| Gene Name |
CPB2
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 3 Target-related Diseases | + | ||||
| 1 | Cerebral ischaemia [ICD-11: 8B1Z] | |||||
| 2 | Cerebral ischaemic stroke [ICD-11: 8B11] | |||||
| 3 | Thrombosis [ICD-11: DB61-GB90] | |||||
| Function |
Cleaves C-terminal arginine or lysine residues from biologically active peptides such as kinins or anaphylatoxins in the circulation thereby regulating their activities. Down- regulates fibrinolysis by removing C-terminal lysine residues from fibrin that has already been partially degraded by plasmin.
Click to Show/Hide
|
|||||
| BioChemical Class |
Peptidase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 3.4.17.20
|
|||||
| Sequence |
MKLCSLAVLVPIVLFCEQHVFAFQSGQVLAALPRTSRQVQVLQNLTTTYEIVLWQPVTAD
LIVKKKQVHFFVNASDVDNVKAHLNVSGIPCSVLLADVEDLIQQQISNDTVSPRASASYY EQYHSLNEIYSWIEFITERHPDMLTKIHIGSSFEKYPLYVLKVSGKEQAAKNAIWIDCGI HAREWISPAFCLWFIGHITQFYGIIGQYTNLLRLVDFYVMPVVNVDGYDYSWKKNRMWRK NRSFYANNHCIGTDLNRNFASKHWCEEGASSSSCSETYCGLYPESEPEVKAVASFLRRNI NQIKAYISMHSYSQHIVFPYSYTRSKSKDHEELSLVASEAVRAIEKISKNTRYTHGHGSE TLYLAPGGGDDWIYDLGIKYSFTIELRDTGTYGFLLPERYIKPTCREAFAAVSKIAWHVI RNV Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 4 Clinical Trial Drugs | + | ||||
| 1 | DS-1040 | Drug Info | Phase 1 | Ischemic stroke | [2] | |
| 2 | SAR-104772 | Drug Info | Phase 1 | Cerebrovascular ischaemia | [3] | |
| 3 | SAR-126119 | Drug Info | Phase 1 | Cerebrovascular ischaemia | [4] | |
| 4 | UK-396082 | Drug Info | Phase 1 | Thrombosis | [5], [6] | |
| Discontinued Drug(s) | [+] 1 Discontinued Drugs | + | ||||
| 1 | AZD-9684 | Drug Info | Discontinued in Phase 2 | Thrombosis | [7] | |
| Mode of Action | [+] 2 Modes of Action | + | ||||
| Modulator | [+] 1 Modulator drugs | + | ||||
| 1 | DS-1040 | Drug Info | [8] | |||
| Inhibitor | [+] 17 Inhibitor drugs | + | ||||
| 1 | SAR-104772 | Drug Info | [9] | |||
| 2 | SAR-126119 | Drug Info | [1] | |||
| 3 | UK-396082 | Drug Info | [10] | |||
| 4 | AZD-9684 | Drug Info | [11] | |||
| 5 | (+/-)-5-amino-2-(mercaptomethyl)pentanoic acid | Drug Info | [12] | |||
| 6 | 2-(3-aminophenyl)-3-mercaptopropanoic acid | Drug Info | [13] | |||
| 7 | 2-(3-guanidinophenyl)-3-mercaptopropanoic acid | Drug Info | [13] | |||
| 8 | 2-(4-benzoylpiperidine-1-carbonyl)benzoic acid | Drug Info | [14] | |||
| 9 | 3-mercapto-2-(piperidin-3-yl)propanoic acid | Drug Info | [13] | |||
| 10 | 3-mercapto-2-(piperidin-4-yl)propanoic acid | Drug Info | [13] | |||
| 11 | 6-Amino-2-(1H-imidazol-4-yl)-hexanoic acid | Drug Info | [15] | |||
| 12 | DL-benzylsuccinic acid | Drug Info | [14] | |||
| 13 | DL-guanidinoethylmercaptosuccinic acid | Drug Info | [14] | |||
| 14 | MN-462 | Drug Info | [16] | |||
| 15 | PMID14640538C3 | Drug Info | [17] | |||
| 16 | PMID19954973C4 | Drug Info | [10] | |||
| 17 | SQ-24798 | Drug Info | [13] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Arginine | Ligand Info | |||||
| Structure Description | Crystal structure of a T325I/T329I/H333Y/H335Q mutant of Thrombin-Activatable Fibrinolysis Inhibitor (TAFI-IIYQ) | PDB:3D68 | ||||
| Method | X-ray diffraction | Resolution | 2.80 Å | Mutation | Yes | [18] |
| PDB Sequence |
AQSGQVLAAL
10 PRTSRQVQVL20 QNLTTTYEIV30 LWQPVTADLI40 VKKKQVHFFV50 NASDVDNVKA 60 HLNVSGIPCS70 VLLADVEDLI80 QQQISNDTVS90 PRASASYYEQ100 YHSLNEIYSW 110 IEFITERHPD120 MLTKIHIGSS130 FEKYPLYVLK140 VSGKEQTAKN150 AIWIDCGIHA 160 REWISPAFCL170 WFIGHITQFY180 GIIGQYTNLL190 RLVDFYVMPV200 VNVDGYDYSW 210 KKNRMWRKNR220 SFYANNHCIG230 TDLNRNFASK240 HWCEEGASSS250 SCSETYCGLY 260 PESEPEVKAV270 ASFLRRNINQ280 IKAYISMHSY290 SQHIVFPYSY300 TRSKSKDHEE 310 LSLVASEAVR320 AIEKISKNIR330 YTYGQGSETL340 YLAPGGGDDW350 IYDLGIKYSF 360 TIELRDTGTY370 GFLLPERYIK380 PTCREAFAAV390 SKIAWHVIRN400 V |
|||||
|
|
||||||
| Ligand Name: (2-Guanidinoethylmercapto)succinic acid | Ligand Info | |||||
| Structure Description | Crystal structure of Thrombin-Activatable Fibrinolysis Inhibitor (TAFI) in complex with 2-guanidino-ethyl-mercaptosuccinic acid (GEMSA) | PDB:3D67 | ||||
| Method | X-ray diffraction | Resolution | 3.40 Å | Mutation | No | [18] |
| PDB Sequence |
AQSGQVLAAL
10 PRTSRQVQVL20 QNLTTTYEIV30 LWQPVTADLI40 VKKKQVHFFV50 NASDVDNVKA 60 HLNVSGIPCS70 VLLADVEDLI80 QQQISNDTVS90 PRASASYYEQ100 YHSLNEIYSW 110 IEFITERHPD120 MLTKIHIGSS130 FEKYPLYVLK140 VSGKEQTAKN150 AIWIDCGIHA 160 REWISPAFCL170 WFIGHITQFY180 GIIGQYTNLL190 RLVDFYVMPV200 VNVDGYDYSW 210 KKNRMWRKNR220 SFYANNHCIG230 TDLNRNFASK240 HWCEEGASSS250 SCSETYCGLY 260 PESEPEVKAV270 ASFLRRNINQ280 IKAYISMHSY290 SQHIVFPYSY300 TRSKSKDHEE 310 LSLVASEAVR320 AIEKTSKNTR330 YTHGHGSETL340 YLAPGGGDDW350 IYDLGIKYSF 360 TIELRDTGTY370 GFLLPERYIK380 PTCREAFAAV390 SKIAWHVIRN400 V |
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
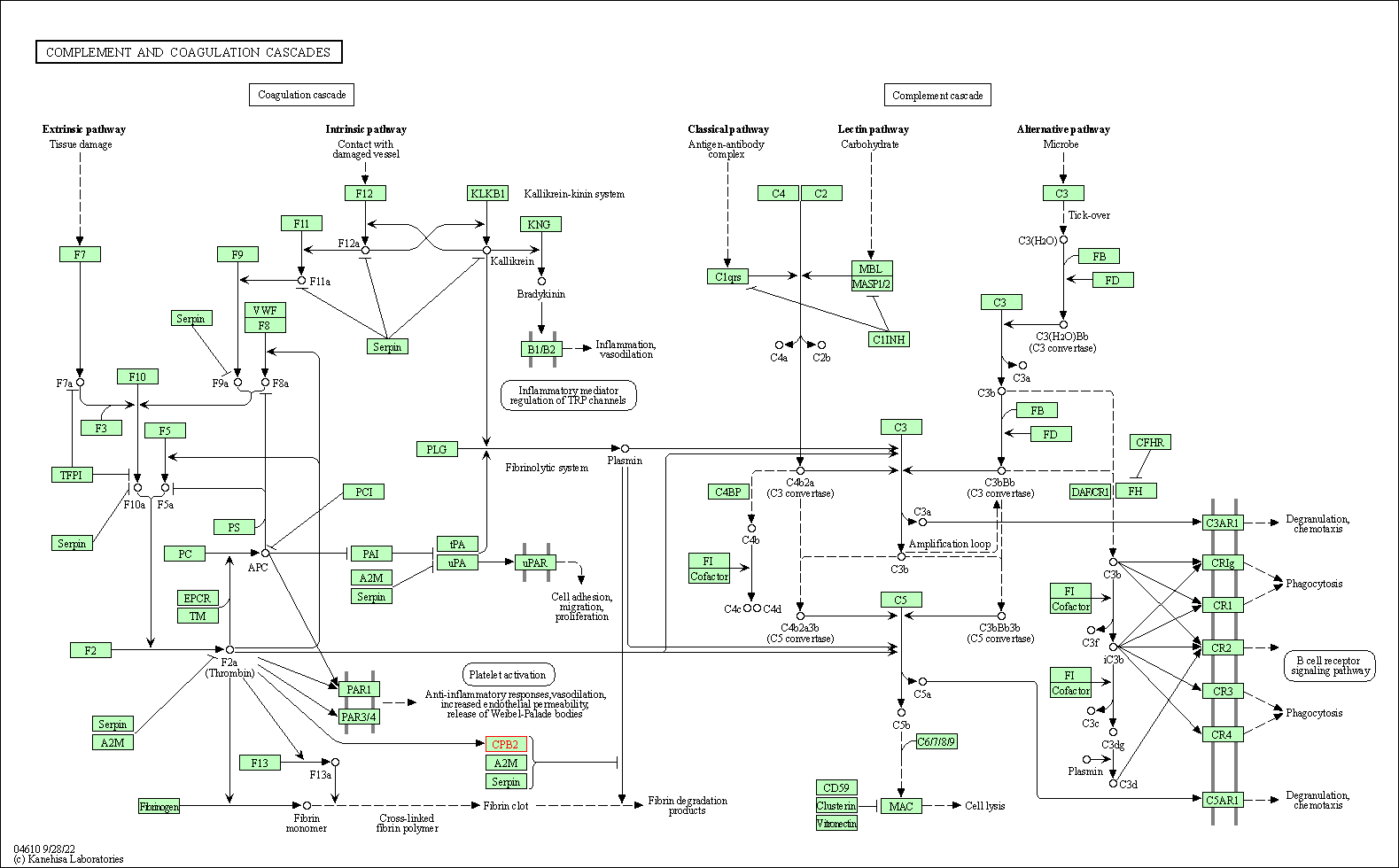
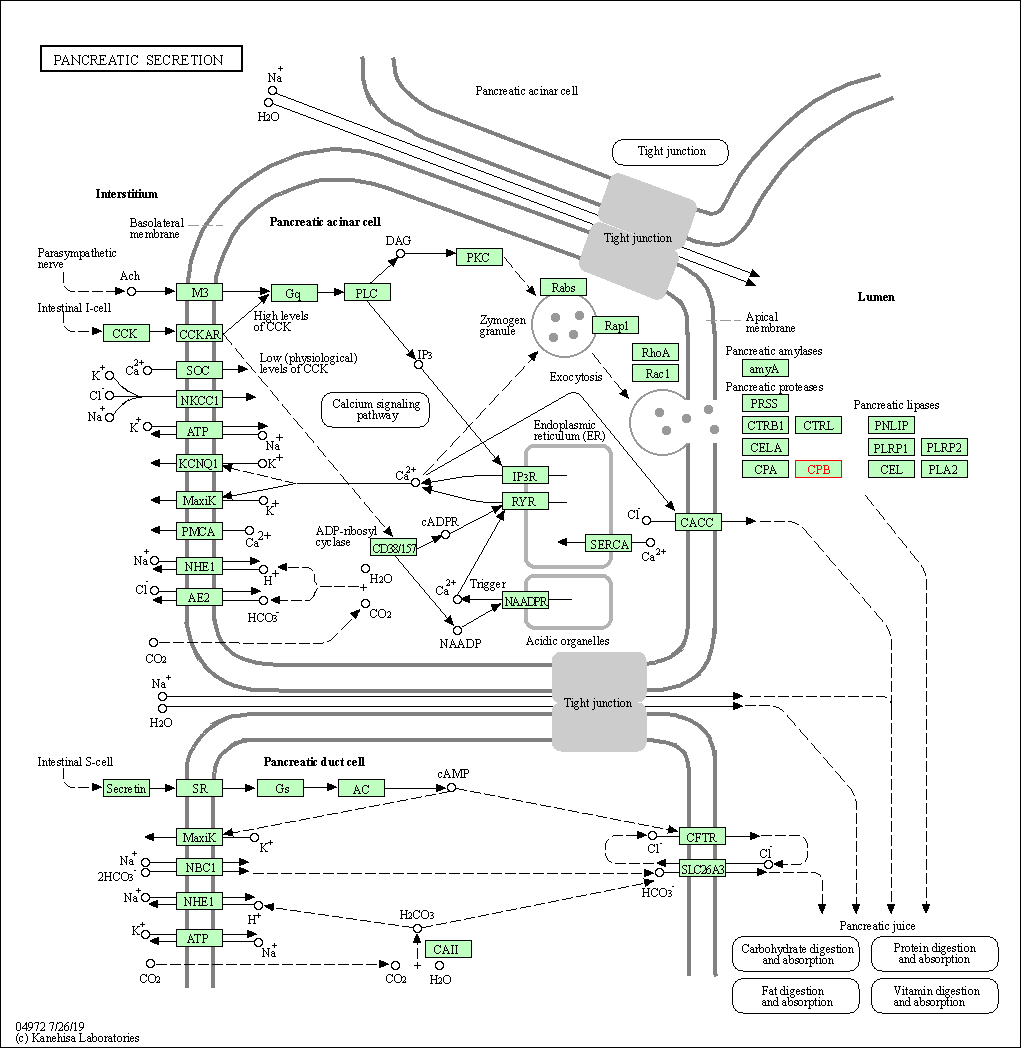
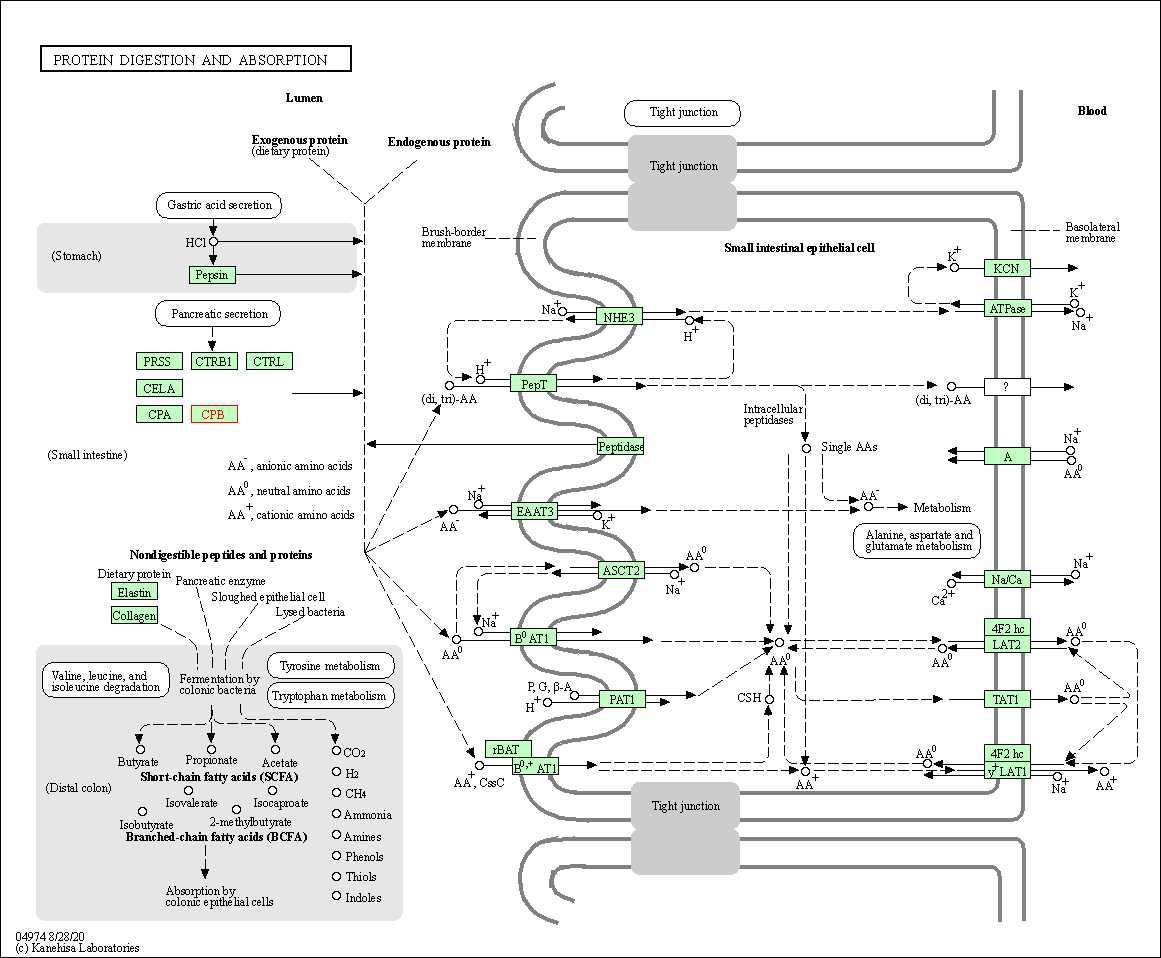
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Complement and coagulation cascades | hsa04610 | Affiliated Target |

|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Pancreatic secretion | hsa04972 | Affiliated Target |

|
| Class: Organismal Systems => Digestive system | Pathway Hierarchy | ||
| Protein digestion and absorption | hsa04974 | Affiliated Target |

|
| Class: Organismal Systems => Digestive system | Pathway Hierarchy | ||
| Degree | 4 | Degree centrality | 4.30E-04 | Betweenness centrality | 2.92E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 1.88E-01 | Radiality | 1.32E+01 | Clustering coefficient | 1.67E-01 |
| Neighborhood connectivity | 1.95E+01 | Topological coefficient | 2.68E-01 | Eccentricity | 13 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Co-Targets | Top | |||||
|---|---|---|---|---|---|---|
| Co-Targets | ||||||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 3 KEGG Pathways | + | ||||
| 1 | Complement and coagulation cascades | |||||
| 2 | Pancreatic secretion | |||||
| 3 | Protein digestion and absorption | |||||
| Panther Pathway | [+] 1 Panther Pathways | + | ||||
| 1 | Plasminogen activating cascade | |||||
| Reactome | [+] 1 Reactome Pathways | + | ||||
| 1 | Metabolism of Angiotensinogen to Angiotensins | |||||
| WikiPathways | [+] 1 WikiPathways | + | ||||
| 1 | Complement and Coagulation Cascades | |||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | TAFIa inhibitors in the treatment of thrombosis. Curr Opin Drug Discov Devel. 2008 Jul;11(4):480-6. | |||||
| REF 2 | ClinicalTrials.gov (NCT02071004) DS1040b/Aspirin Drug/Drug Interaction Study. U.S. National Institutes of Health. | |||||
| REF 3 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800028193) | |||||
| REF 4 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800034720) | |||||
| REF 5 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 8657). | |||||
| REF 6 | ClinicalTrials.gov (NCT01091532) A Study In Healthy People Of Multiple Doses Of UK-396,082 Given By Mouth, To Investigate The Safety, Toleration And Time Course Of Blood Concentration Of UK-396,082 And Its Effects.. U.S. National Institutes of Health. | |||||
| REF 7 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800018349) | |||||
| REF 8 | Clinical pipeline report, company report or official report of Daiichi Sankyo. | |||||
| REF 9 | Carboxypeptidase U (TAFIa): a new drug target for fibrinolytic therapy . Journal of Thrombosis and Haemostasis Volume 7, Issue 12, pages 1962-1971, December 2009. | |||||
| REF 10 | Oxygenated analogues of UK-396082 as inhibitors of activated thrombin activatable fibrinolysis inhibitor. Bioorg Med Chem Lett. 2010 Jan 1;20(1):92-6. | |||||
| REF 11 | Clinical pipeline report, company report or official report of AstraZeneca. | |||||
| REF 12 | Discovery of potent & selective inhibitors of activated thrombin-activatable fibrinolysis inhibitor for the treatment of thrombosis. J Med Chem. 2007 Nov 29;50(24):6095-103. | |||||
| REF 13 | 3-Mercaptopropionic acids as efficacious inhibitors of activated thrombin activatable fibrinolysis inhibitor (TAFIa). Bioorg Med Chem Lett. 2007 Mar 1;17(5):1349-54. | |||||
| REF 14 | A new type of five-membered heterocyclic inhibitors of basic metallocarboxypeptidases. Eur J Med Chem. 2009 Aug;44(8):3266-71. | |||||
| REF 15 | Imidazole acetic acid TAFIa inhibitors: SAR studies centered around the basic P(1)(') group. Bioorg Med Chem Lett. 2004 May 3;14(9):2141-5. | |||||
| REF 16 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Target id: 1594). | |||||
| REF 17 | Synthesis and evaluation of imidazole acetic acid inhibitors of activated thrombin-activatable fibrinolysis inhibitor as novel antithrombotics. J Med Chem. 2003 Dec 4;46(25):5294-7. | |||||
| REF 18 | Crystal structures of TAFI elucidate the inactivation mechanism of activated TAFI: a novel mechanism for enzyme autoregulation. Blood. 2008 Oct 1;112(7):2803-9. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

