Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T94980
|
|||||
| Target Name |
E3 ubiquitin-protein ligase NEDD4 (NEDD4)
|
|||||
| Synonyms |
Neural precursor cell expressed developmentally down-regulated protein 4; NEDD4-1; NEDD-4; KIAA0093; HECT-type E3 ubiquitin transferase NEDD4; Cell proliferation-inducing gene 53 protein
Click to Show/Hide
|
|||||
| Gene Name |
NEDD4
|
|||||
| Target Type |
Literature-reported target
|
[1] | ||||
| Function |
E3 ubiquitin-protein ligase which accepts ubiquitin from an E2 ubiquitin-conjugating enzyme in the form of a thioester and then directly transfers the ubiquitin to targeted substrates. Specifically ubiquitinates 'Lys-63' in target proteins (PubMed:23644597). Involved in the pathway leading to the degradation of VEGFR-2/KDFR, independently of its ubiquitin-ligase activity. Monoubiquitinates IGF1R at multiple sites, thus leading to receptor internalization and degradation in lysosomes. Ubiquitinates FGFR1, leading to receptor internalization and degradation in lysosomes. Promotes ubiquitination of RAPGEF2. According to PubMed:18562292 the direct link between NEDD4 and PTEN regulation through polyubiquitination described in PubMed:17218260 is questionable. Involved in ubiquitination of ERBB4 intracellular domain E4ICD. Involved in the budding of many viruses. Part of a signaling complex composed of NEDD4, RAP2A and TNIK which regulates neuronal dendrite extension and arborization during development. Ubiquitinates TNK2 and regulates EGF-induced degradation of EGFR and TNF2. Ubiquitinates BRAT1 and this ubiquitination is enhanced in the presence of NDFIP1 (PubMed:25631046).
Click to Show/Hide
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.3.2.26
|
|||||
| Sequence |
MAQSLRLHFAARRSNTYPLSETSGDDLDSHVHMCFKRPTRISTSNVVQMKLTPRQTALAP
LIKENVQSQERSSVPSSENVNKKSSCLQISLQPTRYSGYLQSSNVLADSDDASFTCILKD GIYSSAVVDNELNAVNDGHLVSSPAICSGSLSNFSTSDNGSYSSNGSDFGSCASITSGGS YTNSVISDSSSYTFPPSDDTFLGGNLPSDSTSNRSVPNRNTTPCEIFSRSTSTDPFVQDD LEHGLEIMKLPVSRNTKIPLKRYSSLVIFPRSPSTTRPTSPTSLCTLLSKGSYQTSHQFI ISPSEIAHNEDGTSAKGFLSTAVNGLRLSKTICTPGEVRDIRPLHRKGSLQKKIVLSNNT PRQTVCEKSSEGYSCVSVHFTQRKAATLDCETTNGDCKPEMSEIKLNSDSEYIKLMHRTS ACLPSSQNVDCQININGELERPHSQMNKNHGILRRSISLGGAYPNISCLSSLKHNCSKGG PSQLLIKFASGNEGKVDNLSRDSNRDCTNELSNSCKTRDDFLGQVDVPLYPLPTENPRLE RPYTFKDFVLHPRSHKSRVKGYLRLKMTYLPKTSGSEDDNAEQAEELEPGWVVLDQPDAA CHLQQQQEPSPLPPGWEERQDILGRTYYVNHESRRTQWKRPTPQDNLTDAENGNIQLQAQ RAFTTRRQISEETESVDNRESSENWEIIREDEATMYSNQAFPSPPPSSNLDVPTHLAEEL NARLTIFGNSAVSQPASSSNHSSRRGSLQAYTFEEQPTLPVLLPTSSGLPPGWEEKQDER GRSYYVDHNSRTTTWTKPTVQATVETSQLTSSQSSAGPQSQASTSDSGQQVTQPSEIEQG FLPKGWEVRHAPNGRPFFIDHNTKTTTWEDPRLKIPAHLRGKTSLDTSNDLGPLPPGWEE RTHTDGRIFYINHNIKRTQWEDPRLENVAITGPAVPYSRDYKRKYEFFRRKLKKQNDIPN KFEMKLRRATVLEDSYRRIMGVKRADFLKARLWIEFDGEKGLDYGGVAREWFFLISKEMF NPYYGLFEYSATDNYTLQINPNSGLCNEDHLSYFKFIGRVAGMAVYHGKLLDGFFIRPFY KMMLHKPITLHDMESVDSEYYNSLRWILENDPTELDLRFIIDEELFGQTHQHELKNGGSE IVVTNKNKKEYIYLVIQWRFVNRIQKQMAAFKEGFFELIPQDLIKIFDENELELLMCGLG DVDVNDWREHTKYKNGYSANHQVIQWFWKAVLMMDSEKRIRLLQFVTGTSRVPMNGFAEL YGSNGPQSFTVEQWGTPEKLPRAHTCFNRLDLPPYESFEELWDKLQMAIENTQGFDGVD Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Guanidine | Ligand Info | |||||
| Structure Description | Crystal Structure of the Complex of 3rd WW domain of Human Nedd4 and 1st PPXY Motif of ARRDC3 | PDB:4N7H | ||||
| Method | X-ray diffraction | Resolution | 1.70 Å | Mutation | No | [2] |
| PDB Sequence |
GFLPKGWEVR
430 HAPNGRPFFI440 DHNTKTTTWE450 DPR
|
|||||
|
|
||||||
| Ligand Name: methyl (2E)-4-{[(5-methoxy-1,2-dimethyl-1H-indol-3-yl)carbonyl]amino}but-2-enoate | Ligand Info | |||||
| Structure Description | NEDD4 HECT with covalently bound indole-based inhibitor | PDB:5C91 | ||||
| Method | X-ray diffraction | Resolution | 2.44 Å | Mutation | No | [3] |
| PDB Sequence |
SRDYKRKYEF
528 FRRKLKKQND538 IPNKFEMKLR548 RATVLEDSYR558 RIMGVKRADF568 LKARLWIEFD 578 GEKGLDYGGV588 AREWFFLISK598 EMFNPYYGLF608 EYSATDNYTL618 QINPNSGLCN 628 EDHLSYFKFI638 GRVAGMAVYH648 GKLLDGFFIR658 PFYKMMLHKP668 ITLHDMESVD 678 SEYYNSLRWI688 LENDPTELDL698 RFIIDEELFG708 QTHQHELKNG718 GSEIVVTNKN 728 KKEYIYLVIQ738 WRFVNRIQKQ748 MAAFKEGFFE758 LIPQDLIKIF768 DENELELLMC 778 GLGDVDVNDW788 REHTKYKNGY798 SANHQVIQWF808 WKAVLMMDSE818 KRIRLLQFVT 828 GTSRVPMNGF838 AELYGSNGPQ848 SFTVEQWGTP858 EKLPRAHTCF868 NRLDLPPYES 878 FEELWDKLQM888 AIENT
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Ubiquitin mediated proteolysis | hsa04120 | Affiliated Target |
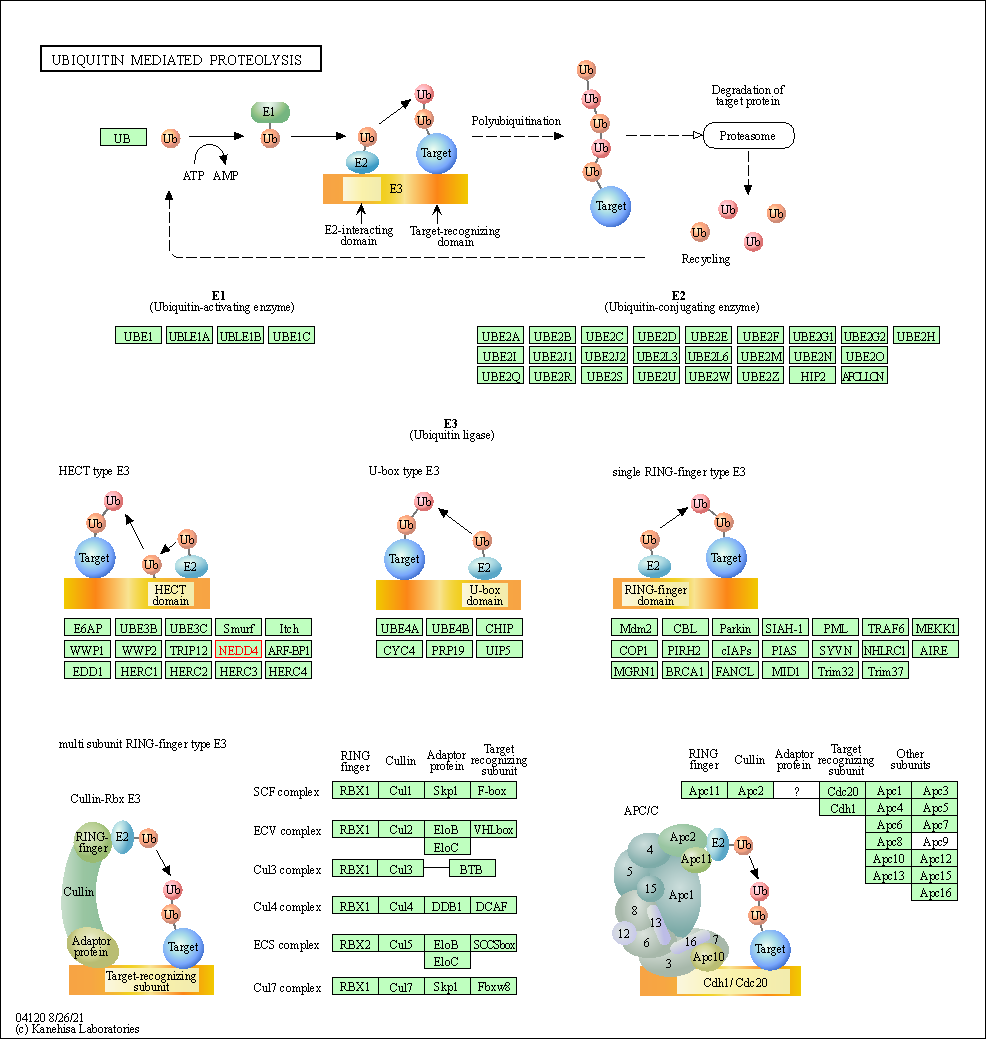
|
| Class: Genetic Information Processing => Folding, sorting and degradation | Pathway Hierarchy | ||
| Endocytosis | hsa04144 | Affiliated Target |
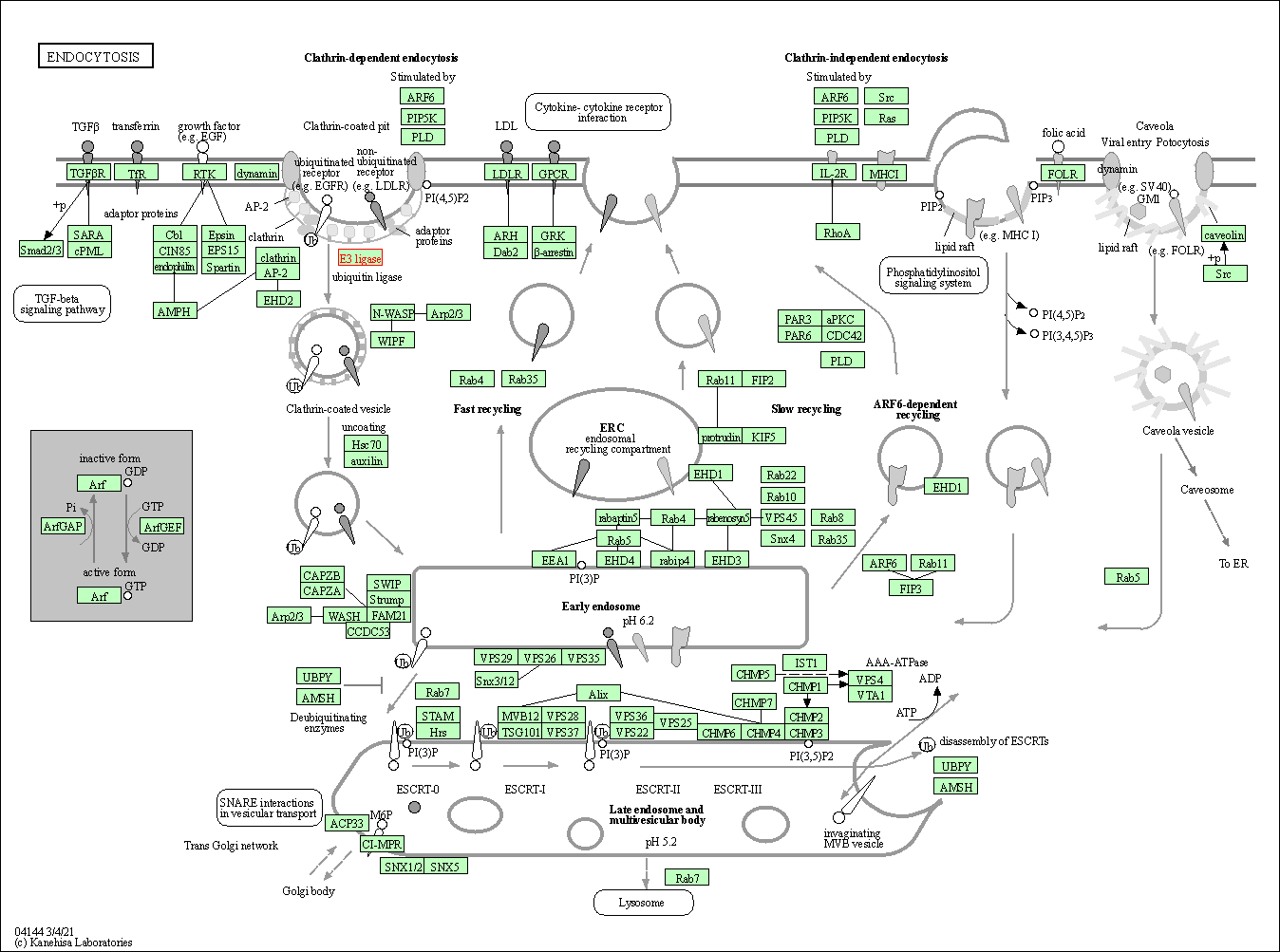
|
| Class: Cellular Processes => Transport and catabolism | Pathway Hierarchy | ||
| Tight junction | hsa04530 | Affiliated Target |
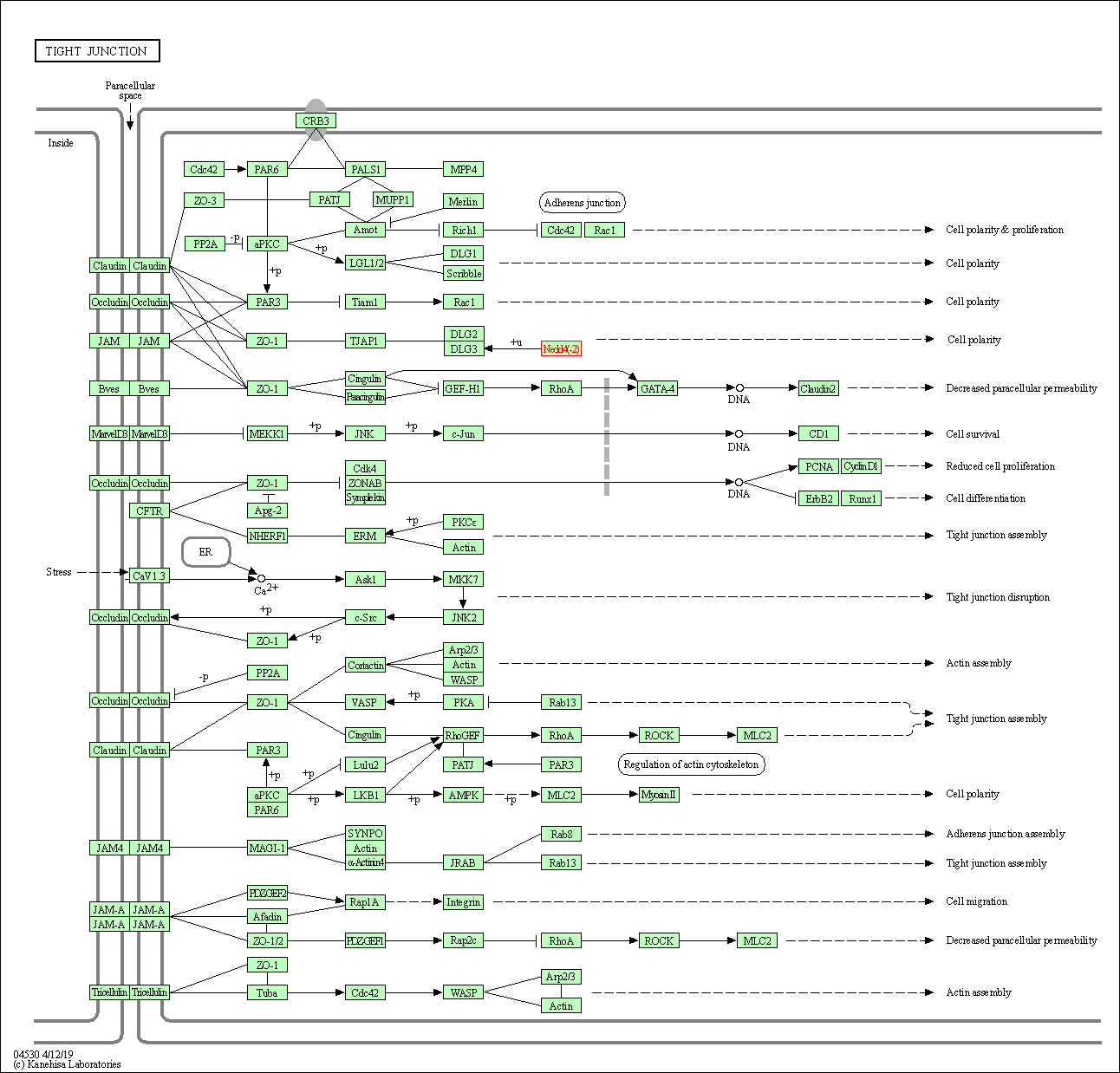
|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
| Degree | 21 | Degree centrality | 2.26E-03 | Betweenness centrality | 1.71E-03 |
|---|---|---|---|---|---|
| Closeness centrality | 2.40E-01 | Radiality | 1.42E+01 | Clustering coefficient | 1.10E-01 |
| Neighborhood connectivity | 4.66E+01 | Topological coefficient | 8.78E-02 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-interacting Proteins | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 4 KEGG Pathways | + | ||||
| 1 | Ubiquitin mediated proteolysis | |||||
| 2 | Endocytosis | |||||
| 3 | Tight junction | |||||
| 4 | Epstein-Barr virus infection | |||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | NEDD4: a promising target for cancer therapy. Curr Cancer Drug Targets. 2014;14(6):549-56. | |||||
| REF 2 | Structural and biochemical basis for ubiquitin ligase recruitment by arrestin-related domain-containing protein-3 (ARRDC3). J Biol Chem. 2014 Feb 21;289(8):4743-52. | |||||
| REF 3 | A Small Molecule That Switches a Ubiquitin Ligase From a Processive to a Distributive Enzymatic Mechanism. J Am Chem Soc. 2015 Oct 7;137(39):12442-5. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

