Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T92517
|
|||||
| Target Name |
Calpain-1 (CAPN1)
|
|||||
| Synonyms |
muCANP; PIG30; Micromolar-calpain; Cell proliferation-inducing gene 30 protein; Calpain-1 large subunit; Calpain-1 catalytic subunit; Calpain mu-type; Calcium-activated neutral proteinase 1; CANPL1; CANP 1
Click to Show/Hide
|
|||||
| Gene Name |
CAPN1
|
|||||
| Target Type |
Patented-recorded target
|
[1] | ||||
| Function |
Calcium-regulated non-lysosomal thiol-protease which catalyzes limited proteolysis of substrates involved in cytoskeletal remodeling and signal transduction.
Click to Show/Hide
|
|||||
| BioChemical Class |
Peptidase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 3.4.22.52
|
|||||
| Sequence |
MSEEIITPVYCTGVSAQVQKQRARELGLGRHENAIKYLGQDYEQLRVRCLQSGTLFRDEA
FPPVPQSLGYKDLGPNSSKTYGIKWKRPTELLSNPQFIVDGATRTDICQGALGDCWLLAA IASLTLNDTLLHRVVPHGQSFQNGYAGIFHFQLWQFGEWVDVVVDDLLPIKDGKLVFVHS AEGNEFWSALLEKAYAKVNGSYEALSGGSTSEGFEDFTGGVTEWYELRKAPSDLYQIILK ALERGSLLGCSIDISSVLDMEAITFKKLVKGHAYSVTGAKQVNYRGQVVSLIRMRNPWGE VEWTGAWSDSSSEWNNVDPYERDQLRVKMEDGEFWMSFRDFMREFTRLEICNLTPDALKS RTIRKWNTTLYEGTWRRGSTAGGCRNYPATFWVNPQFKIRLDETDDPDDYGDRESGCSFV LALMQKHRRRERRFGRDMETIGFAVYEVPPELVGQPAVHLKRDFFLANASRARSEQFINL REVSTRFRLPPGEYVVVPSTFEPNKEGDFVLRFFSEKSAGTVELDDQIQANLPDEQVLSE EEIDENFKALFRQLAGEDMEISVKELRTILNRIISKHKDLRTKGFSLESCRSMVNLMDRD GNGKLGLVEFNILWNRIRNYLSIFRKFDLDKSGSMSAYEMRMAIESAGFKLNKKLYELII TRYSEPDLAVDFDNFVCCLVRLETMFRFFKTLDTDLDGVVTFDLFKWLQLTMFA Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| HIT2.0 ID | T26TBJ | |||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: N-[(Benzyloxy)carbonyl]leucyl-N~1~-[3-Fluoro-1-(4-Hydroxybenzyl)-2-Oxopropyl]leucinamide | Ligand Info | |||||
| Structure Description | Human calpain protease core inhibited by ZLLYCH2F | PDB:1ZCM | ||||
| Method | X-ray diffraction | Resolution | 2.00 Å | Mutation | Yes | [2] |
| PDB Sequence |
NAIKYLGQDY
42 EQLRVRCLQS52 GTLFRDEAFP62 PVPQSLGYKD72 LGPNSSKTYG82 IKWKRPTELL 92 SNPQFIVDGA102 TRTDICQGAL112 GDCWLLAAIA122 SLTLNDTLLH132 RVVPHGQSFQ 142 NGYAGIFHFQ152 LWQFGEWVDV162 VVDDLLPIKD172 GKLVFVHSAE182 GNEFWSALLE 192 KAYAKVNGSY202 EALSGGSTSE212 AFEDFTGGVT222 EWYELRKAPS232 DLYQIILKAL 242 ERGSLLGCSI252 DISSVLDMEA262 ITFKKLVKGH272 AYSVTGAKQV282 NYRGQVVSLI 292 RMRNPWGEVE302 WTGAWSDSSS312 EWNNVDPYER322 DQLRVKMEDG332 EFWMSFRDFM 342 REFTRLEICN352 L
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Protein processing in endoplasmic reticulum | hsa04141 | Affiliated Target |
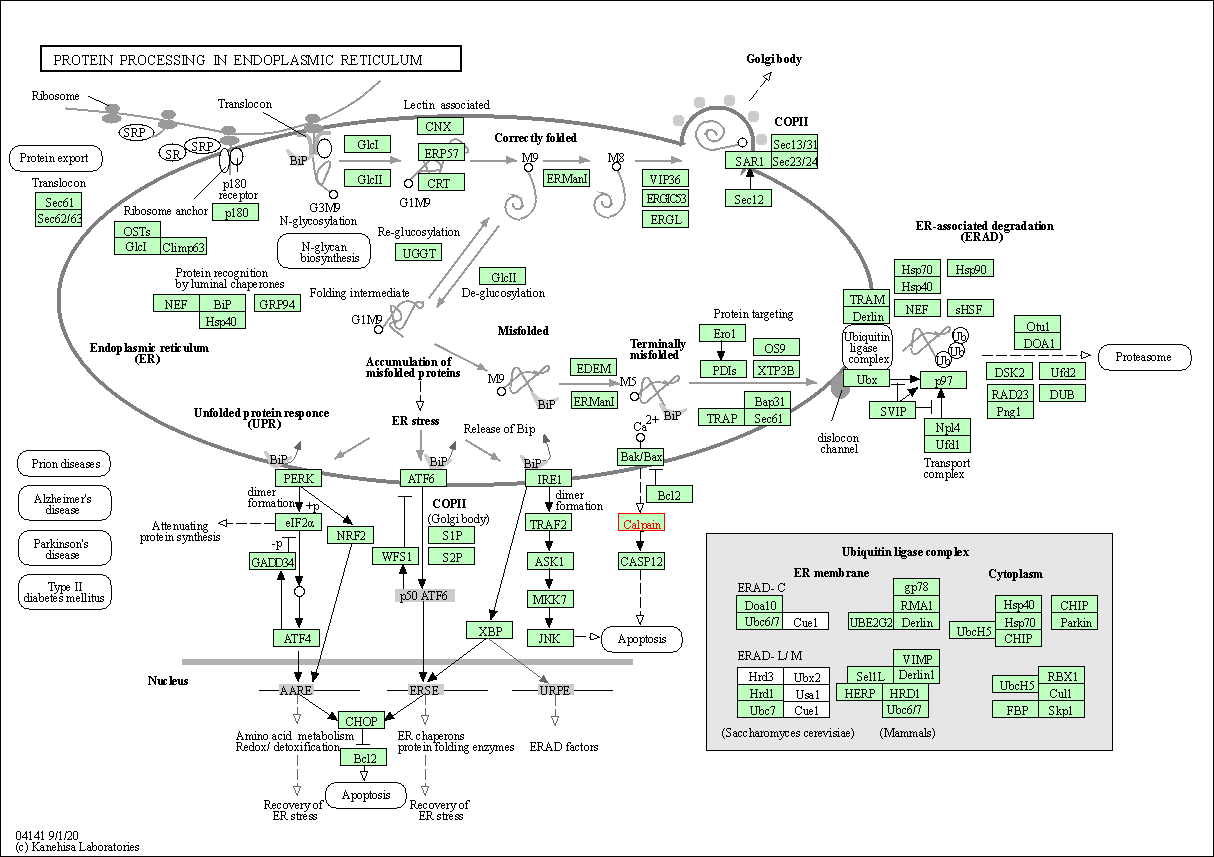
|
| Class: Genetic Information Processing => Folding, sorting and degradation | Pathway Hierarchy | ||
| Apoptosis | hsa04210 | Affiliated Target |
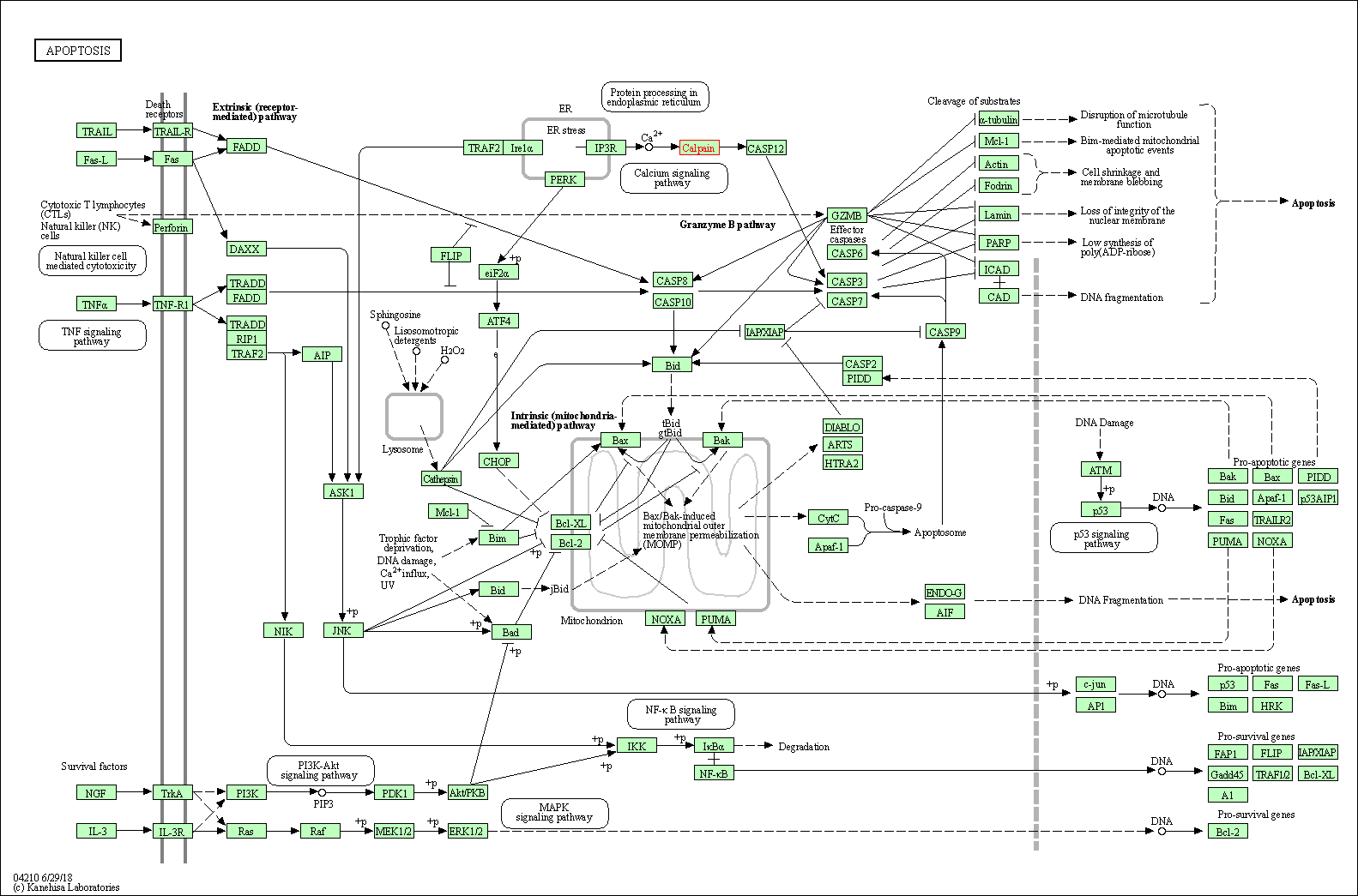
|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Necroptosis | hsa04217 | Affiliated Target |
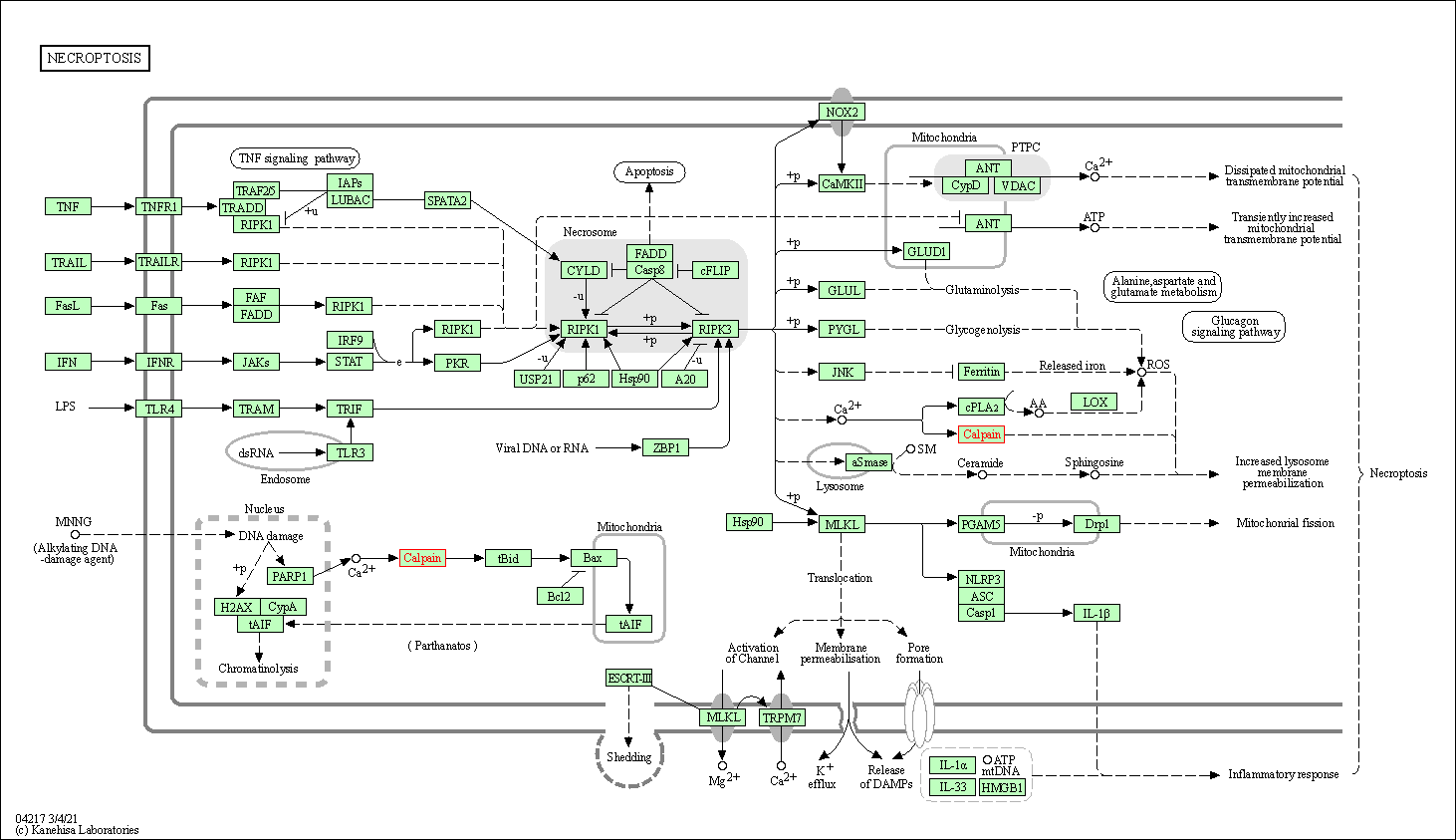
|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Cellular senescence | hsa04218 | Affiliated Target |
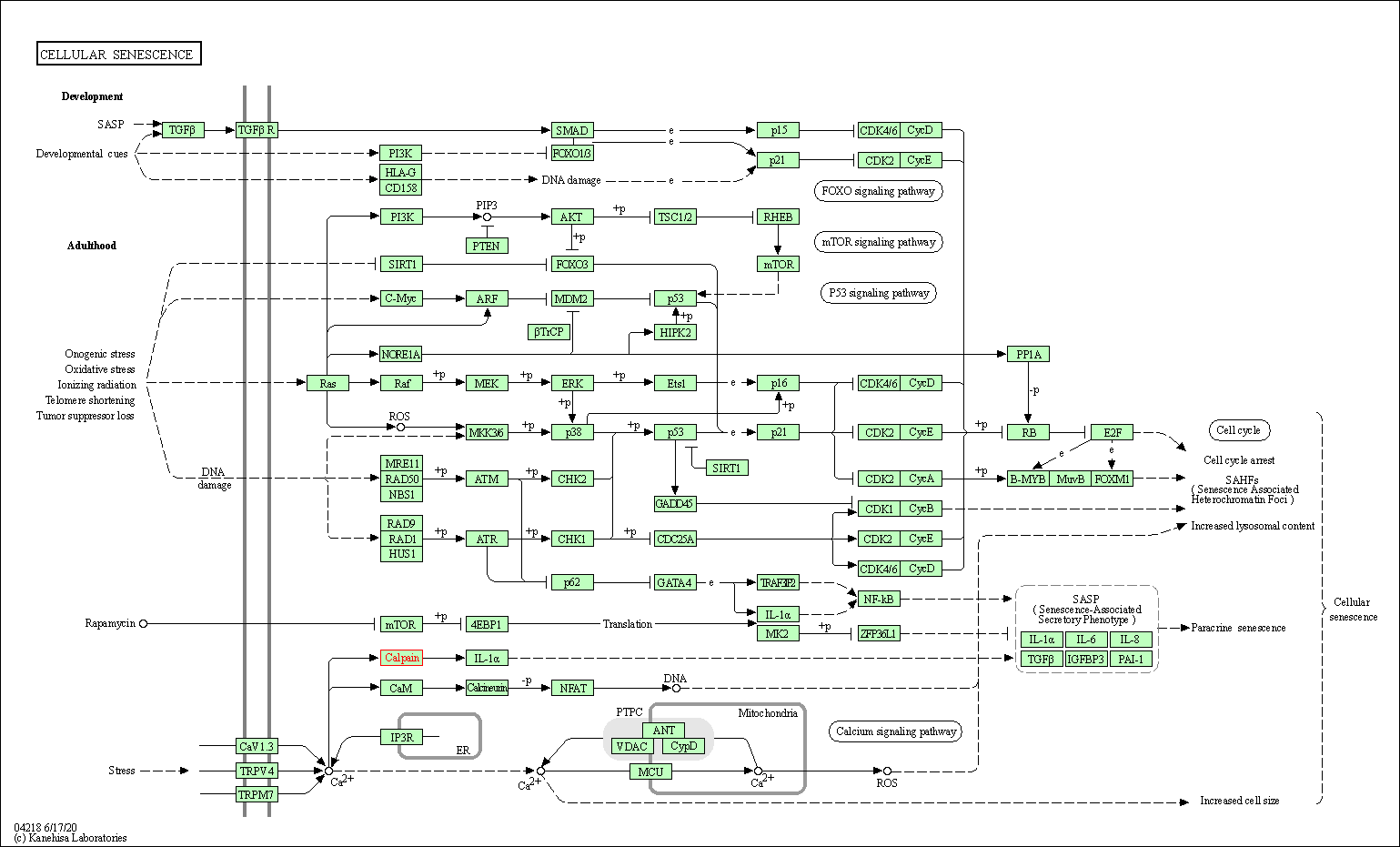
|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Degree | 4 | Degree centrality | 4.30E-04 | Betweenness centrality | 8.58E-05 |
|---|---|---|---|---|---|
| Closeness centrality | 2.01E-01 | Radiality | 1.35E+01 | Clustering coefficient | 1.67E-01 |
| Neighborhood connectivity | 1.08E+01 | Topological coefficient | 2.70E-01 | Eccentricity | 13 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-interacting Proteins | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 5 KEGG Pathways | + | ||||
| 1 | Protein processing in endoplasmic reticulum | |||||
| 2 | Apoptosis | |||||
| 3 | Necroptosis | |||||
| 4 | Cellular senescence | |||||
| 5 | Alzheimer disease | |||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | An updated patent review of calpain inhibitors (2012 - 2014).Expert Opin Ther Pat. 2015 Jan;25(1):17-31. | |||||
| REF 2 | Molecular mode of action of a covalently inhibiting peptidomimetic on the human calpain protease core. Biochemistry. 2006 Jan 24;45(3):701-8. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

