Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T88569
|
|||||
| Target Name |
Mutated oxalosuccinate decarboxylase (mIDH1)
|
|||||
| Synonyms |
PICD (mutated); Oxalosuccinate decarboxylase (mutated); NADP(+)-specific ICDH (mutated); Isocitrate dehydrogenase [NADP] cytoplasmic (mutated); IDP (mutated); IDH (mutated); Cytosolic NADP-isocitrate dehydrogenase (mutated)
Click to Show/Hide
|
|||||
| Gene Name |
IDH1
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 2 Target-related Diseases | + | ||||
| 1 | Acute myeloid leukaemia [ICD-11: 2A60] | |||||
| 2 | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||||
| Function |
Catalyses the NADPH-dependent reduction of alpha-ketoglutarate to R(-)-2-hydroxyglutarate (2HG).
Click to Show/Hide
|
|||||
| BioChemical Class |
Short-chain dehydrogenases reductase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 1.1.1.42
|
|||||
| Sequence |
MSKKISGGSVVEMQGDEMTRIIWELIKEKLIFPYVELDLHSYDLGIENRDATNDQVTKDA
AEAIKKHNVGVKCATITPDEKRVEEFKLKQMWKSPNGTIRNILGGTVFREAIICKNIPRL VSGWVKPIIIGRHAYGDQYRATDFVVPGPGKVEITYTPSDGTQKVTYLVHNFEEGGGVAM GMYNQDKSIEDFAHSSFQMALSKGWPLYLSTKNTILKKYDGRFKDIFQEIYDKQYKSQFE AQKIWYEHRLIDDMVAQAMKSEGGFIWACKNYDGDVQSDSVAQGYGSLGMMTSVLVCPDG KTVEAEAAHGTVTRHYRMYQKGQETSTNPIASIFAWTRGLAHRAKLDNNKELAFFANALE EVSIETIEAGFMTKDLAACIKGLPNVQRSDYLNTFEFMDKLGENLKIKLAQAKL Click to Show/Hide
|
|||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 1 Clinical Trial Drugs | + | ||||
| 1 | BAY1436032 | Drug Info | Phase 1 | Acute myeloid leukaemia | [2] | |
| Mode of Action | [+] 1 Modes of Action | + | ||||
| Inhibitor | [+] 1 Inhibitor drugs | + | ||||
| 1 | BAY1436032 | Drug Info | [2] | |||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: AG-881 | Ligand Info | |||||
| Structure Description | Crystal Structures of IDH1 R132H in complex with AG-881 | PDB:6ADG | ||||
| Method | X-ray diffraction | Resolution | 3.00 Å | Mutation | Yes | [3] |
| PDB Sequence |
KKISGGSVVE
12 MQGDEMTRII22 WELIKEKLIF32 PYVELDLHSY42 DLGIENRDAT52 NDQVTKDAAE 62 AIKKHNVGVK72 CATITPDEKR82 VEEFKLKQMW92 KSPNGTIRNI102 LGGTVFREAI 112 ICKNIPRLVS122 GWVKPIIIGH132 HAYGDQYRAT142 DFVVPGPGKV152 EITYTPSDGT 162 QKVTYLVHNF172 EEGGGVAMGM182 YNQDKSIEDF192 AHSSFQMALS202 KGWPLYLSTK 212 NTILKKYDGR222 FKDIFQEIYD232 KQYKSQFEAQ242 KIWYEHRLID252 DMVAQAMKSE 262 GGFIWACKNY272 DGDVQSDSVA282 QGYGSLGMMT292 SVLVCPDGKT302 VEAEAAHGTV 312 TRHYRMYQKG322 QETSTNPIAS332 IFAWTRGLAH342 RAKLDNNKEL352 AFFANALEEV 362 SIETIEAGFM372 TKDLAACIKG382 LPNVQRSDYL392 NTFEFMDKLG402 ENLKIKLAQA 412 KL
|
|||||
|
|
||||||
| Ligand Name: FT-2102 | Ligand Info | |||||
| Structure Description | Crystal structure of IDH1 R132H mutant in complex with FT-2102 | PDB:6U4J | ||||
| Method | X-ray diffraction | Resolution | 2.11 Å | Mutation | Yes | [4] |
| PDB Sequence |
KKISGGSVVE
12 MQGDEMTRII22 WELIKEKLIF32 PYVELDLHSY42 DLGIENRDAT52 NDQVTKDAAE 62 AIKKHNVGVK72 CATITPDEKR82 VEEFKLKQMW92 KSPNGTIRNI102 LGGTVFREAI 112 ICKNIPRLVS122 GWVKPIIIGH132 HAYGDQYRAT142 DFVVPGPGKV152 EITYTPSDGT 162 QKVTYLVHNF172 EEGGGVAMGM182 YNQDKSIEDF192 AHSSFQMALS202 KGWPLYLSTK 212 NTILKKYDGR222 FKDIFQEIYD232 KQYKSQFEAQ242 KIWYEHRLID252 DMVAQAMKSE 262 GGFIWACKNY272 DGDVQSDSVA282 QGYGSLGMMT292 SVLVCPDGKT302 VEAEAAHGTV 312 TRHYRMYQKG322 QETSTNPIAS332 IFAWTRGLAH342 RAKLDNNKEL352 AFFANALEEV 362 SIETIEAGFM372 TKDLAACIKG382 LPNVQRSDYL392 NTFEFMDKLG402 ENLKIKLAQA 412 KLSLE
|
|||||
|
|
ARG109
2.897
GLU110
3.994
ALA111
3.540
ILE113
3.639
ARG119
3.942
LEU120
3.050
VAL121
4.298
TRP124
3.684
LYS126
4.845
PRO127
3.440
ILE128
2.915
|
|||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Human Pathway Affiliation
|
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Citrate cycle (TCA cycle) | hsa00020 | Affiliated Target |
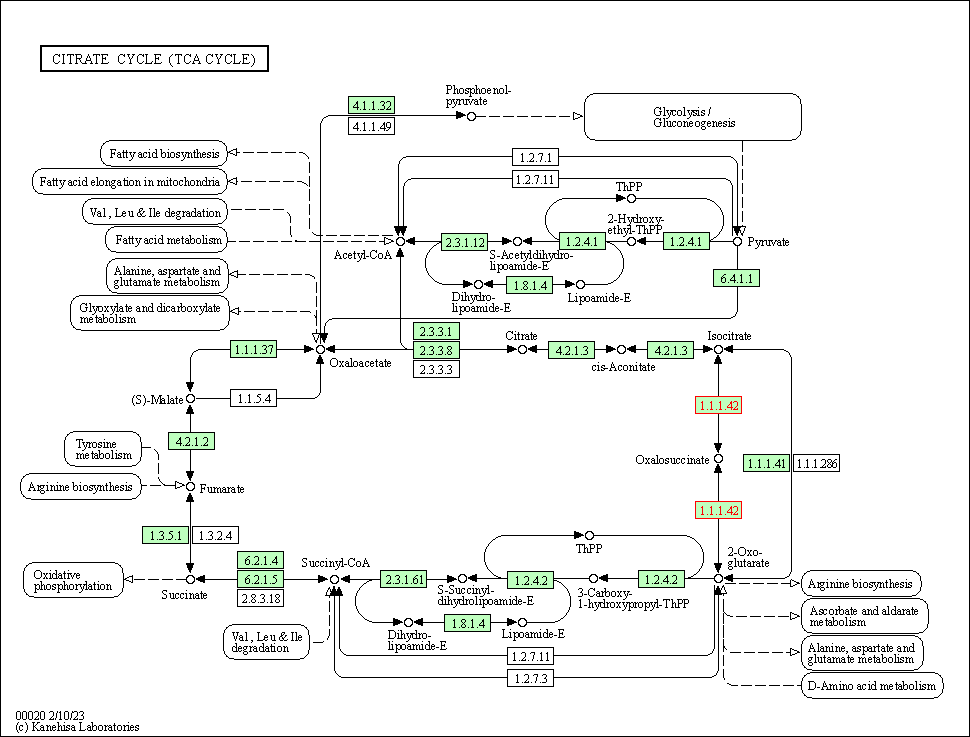
|
| Class: Metabolism => Carbohydrate metabolism | Pathway Hierarchy | ||
| Glutathione metabolism | hsa00480 | Affiliated Target |
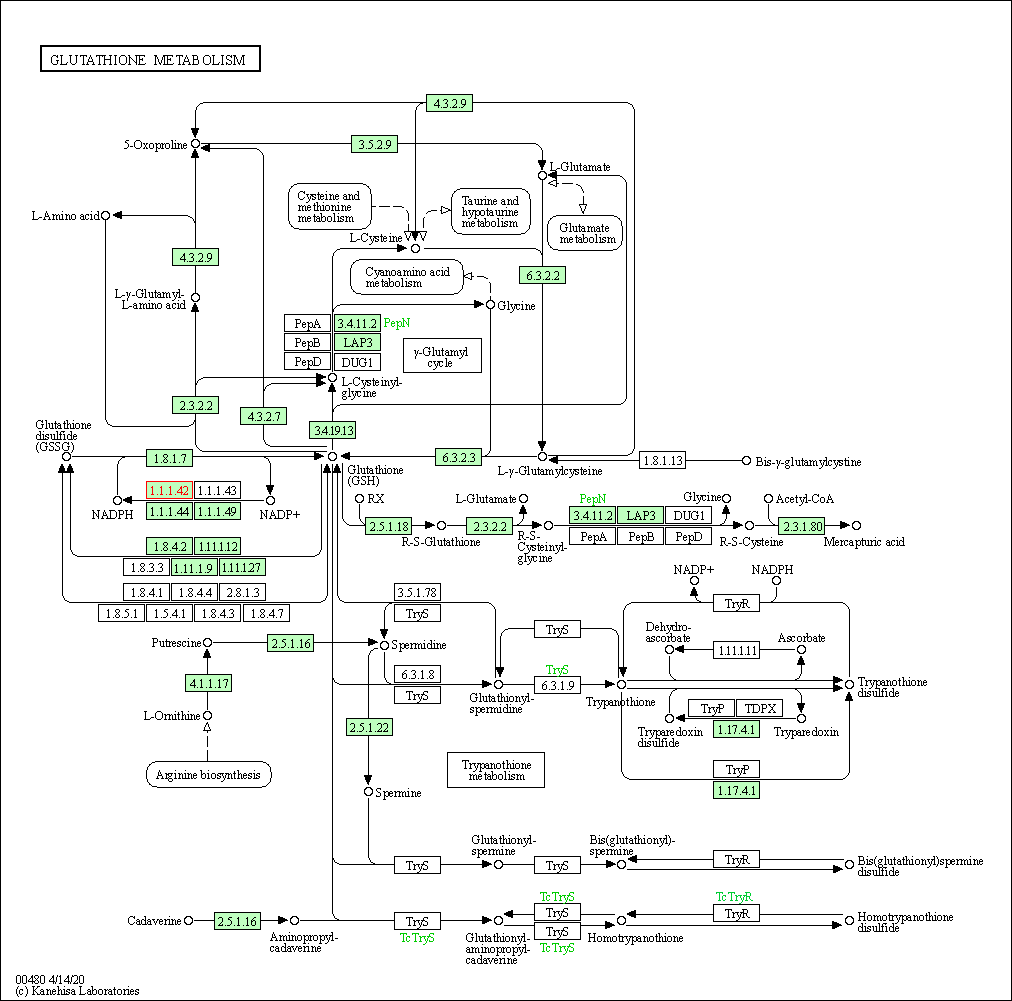
|
| Class: Metabolism => Metabolism of other amino acids | Pathway Hierarchy | ||
| Peroxisome | hsa04146 | Affiliated Target |
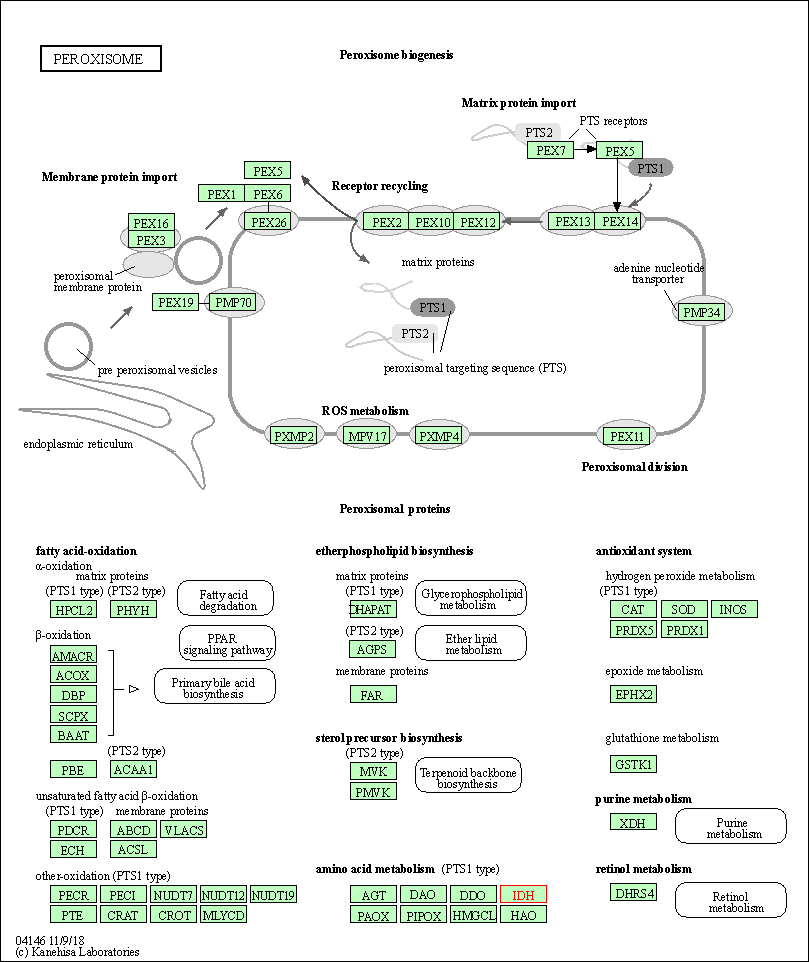
|
| Class: Cellular Processes => Transport and catabolism | Pathway Hierarchy | ||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 2 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 3 | Crystal structures of pan-IDH inhibitor AG-881 in complex with mutant human IDH1 and IDH2. Biochem Biophys Res Commun. 2018 Sep 18;503(4):2912-2917. | |||||
| REF 4 | Structure-Based Design and Identification of FT-2102 (Olutasidenib), a Potent Mutant-Selective IDH1 Inhibitor. J Med Chem. 2020 Feb 27;63(4):1612-1623. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

