Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T73134
|
|||||
| Target Name |
Beta-galactosidase (GLB1)
|
|||||
| Synonyms |
Lactase; Elastin receptor 1; ELNR1; Acid beta-galactosidase
Click to Show/Hide
|
|||||
| Gene Name |
GLB1
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Lysosomal disease [ICD-11: 5C56] | |||||
| Function |
Isoform 1: Cleaves beta-linked terminal galactosyl residues from gangliosides, glycoproteins, and glycosaminoglycans.
Click to Show/Hide
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 3.2.1.23
|
|||||
| Sequence |
MPGFLVRILPLLLVLLLLGPTRGLRNATQRMFEIDYSRDSFLKDGQPFRYISGSIHYSRV
PRFYWKDRLLKMKMAGLNAIQTYVPWNFHEPWPGQYQFSEDHDVEYFLRLAHELGLLVIL RPGPYICAEWEMGGLPAWLLEKESILLRSSDPDYLAAVDKWLGVLLPKMKPLLYQNGGPV ITVQVENEYGSYFACDFDYLRFLQKRFRHHLGDDVVLFTTDGAHKTFLKCGALQGLYTTV DFGTGSNITDAFLSQRKCEPKGPLINSEFYTGWLDHWGQPHSTIKTEAVASSLYDILARG ASVNLYMFIGGTNFAYWNGANSPYAAQPTSYDYDAPLSEAGDLTEKYFALRNIIQKFEKV PEGPIPPSTPKFAYGKVTLEKLKTVGAALDILCPSGPIKSLYPLTFIQVKQHYGFVLYRT TLPQDCSNPAPLSSPLNGVHDRAYVAVDGIPQGVLERNNVITLNITGKAGATLDLLVENM GRVNYGAYINDFKGLVSNLTLSSNILTDWTIFPLDTEDAVRSHLGGWGHRDSGHHDEAWA HNSSNYTLPAFYMGNFSIPSGIPDLPQDTFIQFPGWTKGQVWINGFNLGRYWPARGPQLT LFVPQHILMTSAPNTITVLELEWAPCSSDDPELCAVTFVDRPVIGSSVTYDHPSKPVEKR LMPPPPQKNKDSWLDHV Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| HIT2.0 ID | T19JEU | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 3 Clinical Trial Drugs | + | ||||
| 1 | AXO-AAV-GM1 | Drug Info | Phase 1/2 | GM1 gangliosidosis | [2] | |
| 2 | LYS-GM-101 | Drug Info | Phase 1/2 | GM1 gangliosidosis | [3] | |
| 3 | PBGM01 | Drug Info | Phase 1/2 | GM1 gangliosidosis | [4] | |
| Preclinical Drug(s) | [+] 2 Preclinical Drugs | + | ||||
| 1 | 5N,6S-(N'-butyliminomethylidene)-6-thio-1-deoxygalactonojirimycin | Drug Info | Preclinical | GM1 gangliosidosis | [5] | |
| 2 | N-Octyl-4-epi-beta-valienamine | Drug Info | Preclinical | GM1 gangliosidosis | [5] | |
| Mode of Action | [+] 1 Modes of Action | + | ||||
| Inhibitor | [+] 2 Inhibitor drugs | + | ||||
| 1 | 5N,6S-(N'-butyliminomethylidene)-6-thio-1-deoxygalactonojirimycin | Drug Info | [7] | |||
| 2 | N-Octyl-4-epi-beta-valienamine | Drug Info | [8] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Migalastat | Ligand Info | |||||
| Structure Description | Crystal structure of human beta-galactosidase in complex with 1-deoxygalactonojirimycin | PDB:3THD | ||||
| Method | X-ray diffraction | Resolution | 1.79 Å | Mutation | No | [9] |
| PDB Sequence |
QRMFEIDYSR
38 DSFLKDGQPF48 RYISGSIHYS58 RVPRFYWKDR68 LLKMKMAGLN78 AIQTYVPWNF 88 HEPWPGQYQF98 SEDHDVEYFL108 RLAHELGLLV118 ILRPGPYICA128 EWEMGGLPAW 138 LLEKESILLR148 SSDPDYLAAV158 DKWLGVLLPK168 MKPLLYQNGG178 PVITVQVENE 188 YGSYFACDFD198 YLRFLQKRFR208 HHLGDDVVLF218 TTDGAHKTFL228 KCGALQGLYT 238 TVDFGTGSNI248 TDAFLSQRKC258 EPKGPLINSE268 FYTGWLDHWG278 QPHSTIKTEA 288 VASSLYDILA298 RGASVNLYMF308 IGGTNFAYWN318 GANSPYAAQP328 TSYDYDAPLS 338 EAGDLTEKYF348 ALRNIIQKFE358 KVPEGPIPPS368 TPKFAYGKVT378 LEKLKTVGAA 388 LDILCPSGPI398 KSLYPLTFIQ408 VKQHYGFVLY418 RTTLPQDCSN428 PAPLSSPLNG 438 VHDRAYVAVD448 GIPQGVLERN458 NVITLNITGK468 AGATLDLLVE478 NMGRVNYGAY 488 INDFKGLVSN498 LTLSSNILTD508 WTIFPLDTED518 AVRSHLGGWG528 HRNYTLPAFY 552 MGNFSIPSGI562 PDLPQDTFIQ572 FPGWTKGQVW582 INGFNLGRYW592 PARGPQLTLF 602 VPQHILMTSA612 PNTITVLELE622 WAPCSSDDPE632 LCAVTFVDRP642 VIGSS |
|||||
|
|
||||||
| Ligand Name: N-Octyl-4-epi-beta-valienamine | Ligand Info | |||||
| Structure Description | Crystal structure of human beta-galactosidase in complex with NOEV | PDB:3WEZ | ||||
| Method | X-ray diffraction | Resolution | 2.11 Å | Mutation | No | [10] |
| PDB Sequence |
QRMFEIDYSR
38 DSFLKDGQPF48 RYISGSIHYS58 RVPRFYWKDR68 LLKMKMAGLN78 AIQTYVPWNF 88 HEPWPGQYQF98 SEDHDVEYFL108 RLAHELGLLV118 ILRPGPYICA128 EWEMGGLPAW 138 LLEKESILLR148 SSDPDYLAAV158 DKWLGVLLPK168 MKPLLYQNGG178 PVITVQVENE 188 YGSYFACDFD198 YLRFLQKRFR208 HHLGDDVVLF218 TTDGAHKTFL228 KCGALQGLYT 238 TVDFGTGSNI248 TDAFLSQRKC258 EPKGPLINSE268 FYTGWLDHWG278 QPHSTIKTEA 288 VASSLYDILA298 RGASVNLYMF308 IGGTNFAYWN318 GANSPYAAQP328 TSYDYDAPLS 338 EAGDLTEKYF348 ALRNIIQKFE358 KVPEGPIPPS368 TPKFAYGKVT378 LEKLKTVGAA 388 LDILCPSGPI398 KSLYPLTFIQ408 VKQHYGFVLY418 RTTLPQDCSN428 PAPLSSPLNG 438 VHDRAYVAVD448 GIPQGVLERN458 NVITLNITGK468 AGATLDLLVE478 NMGRVNYGAY 488 INDFKGLVSN498 LTLSSNILTD508 WTIFPLDTED518 AVRSHLGGWG528 HRNYTLPAFY 552 MGNFSIPSGI562 PDLPQDTFIQ572 FPGWTKGQVW582 INGFNLGRYW592 PARGPQLTLF 602 VPQHILMTSA612 PNTITVLELE622 WAPCSSDDPE632 LCAVTFVDRP642 VIGSS |
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
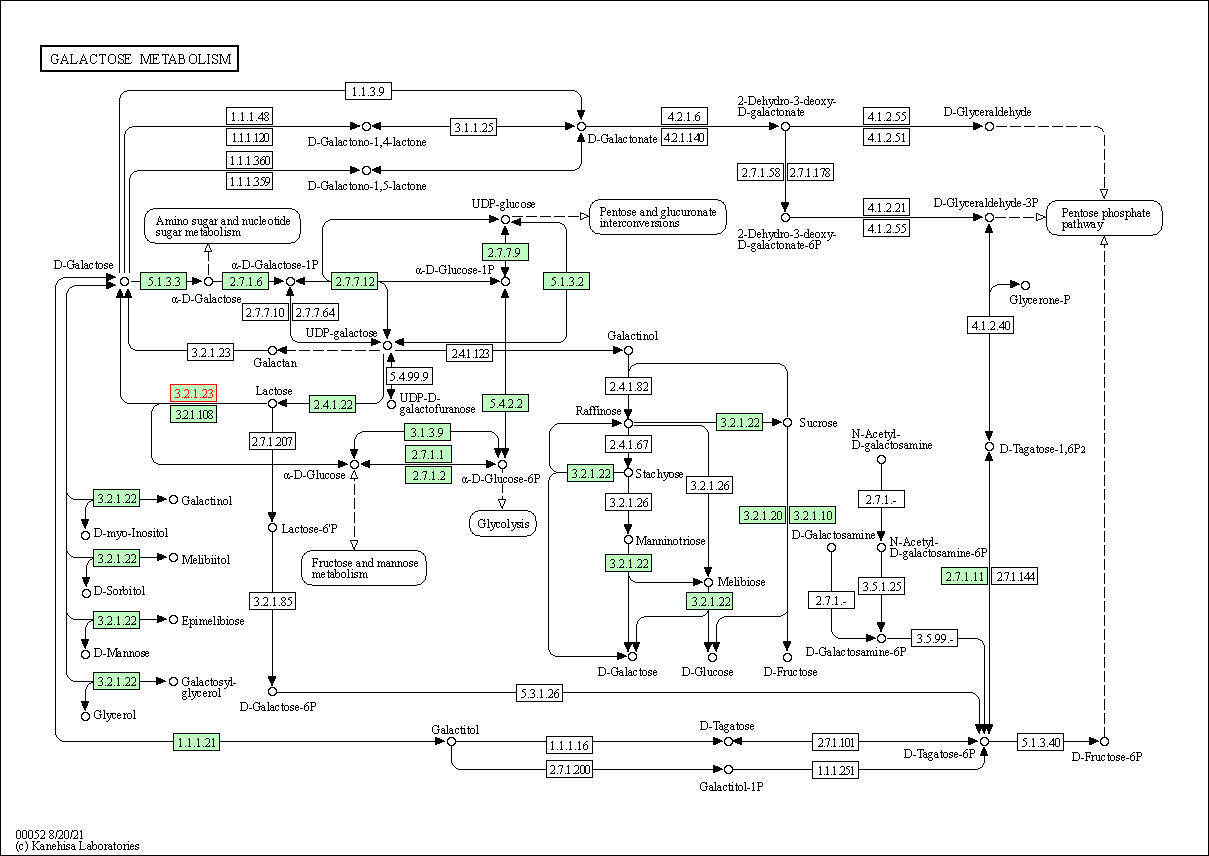
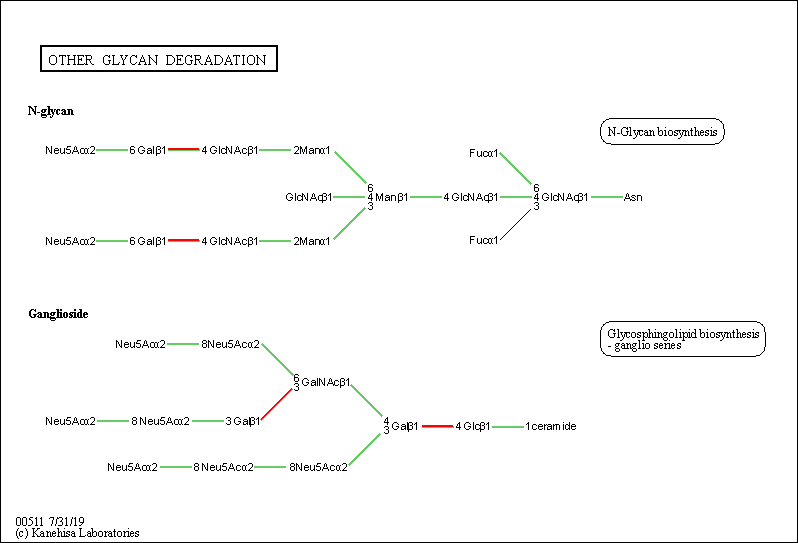
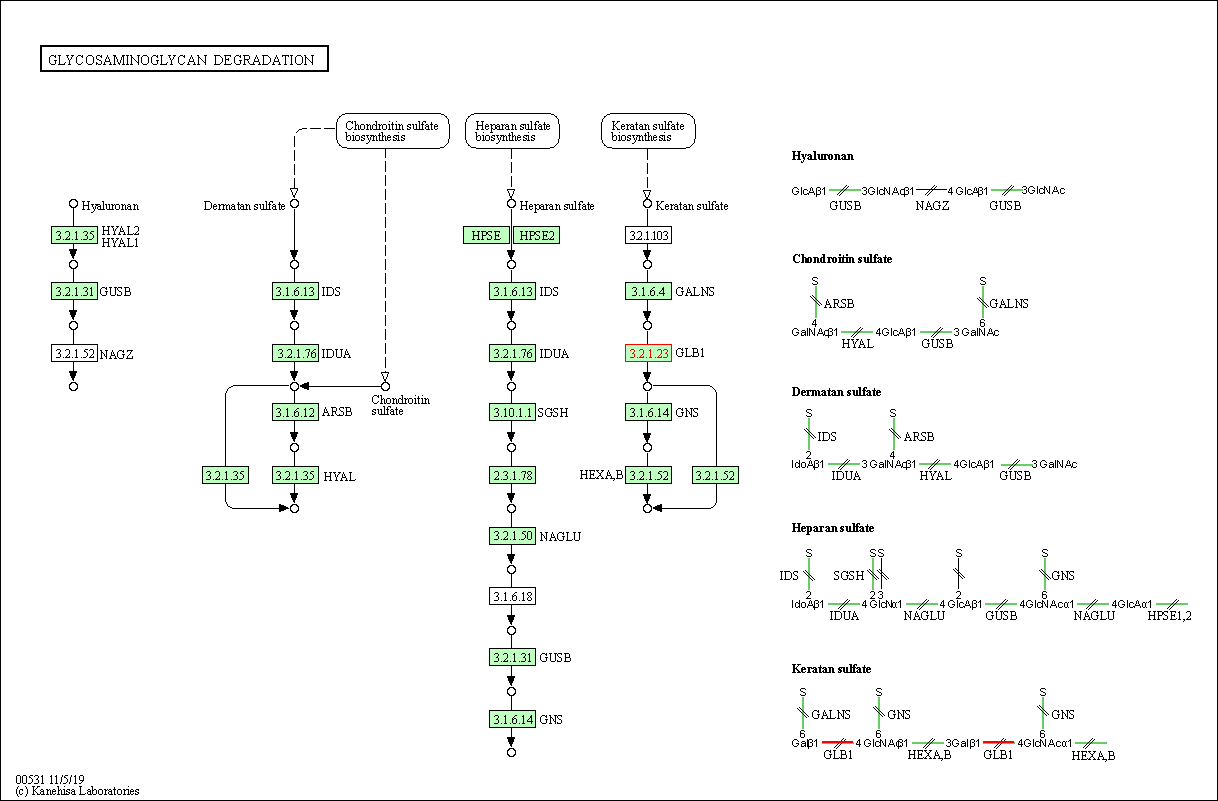

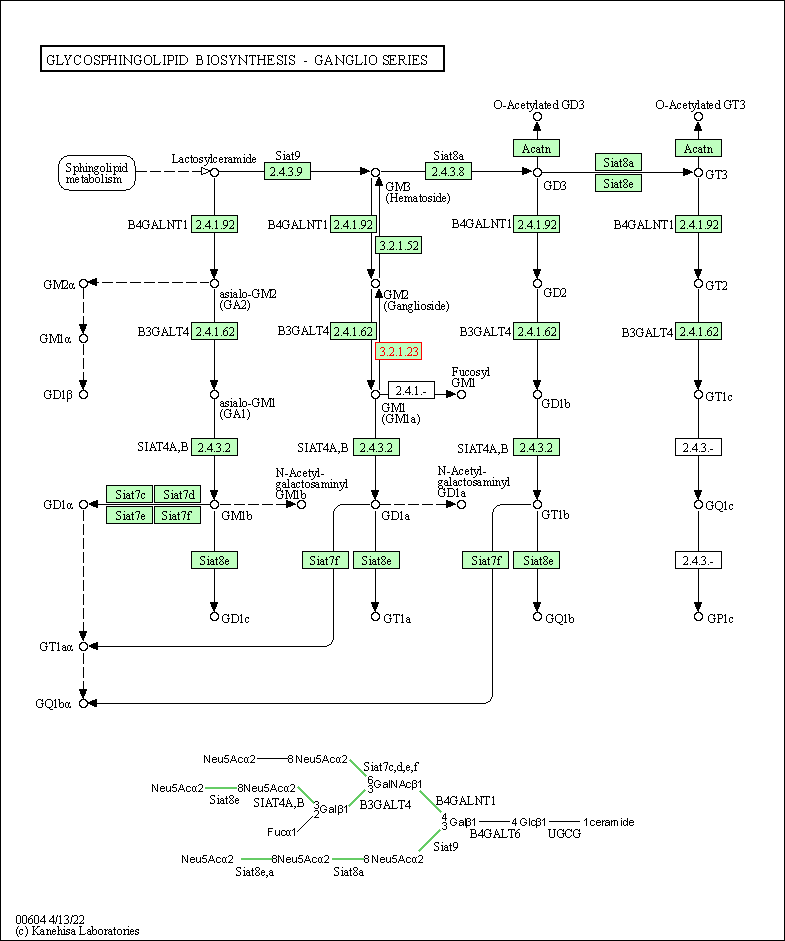
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Galactose metabolism | hsa00052 | Affiliated Target |

|
| Class: Metabolism => Carbohydrate metabolism | Pathway Hierarchy | ||
| Other glycan degradation | hsa00511 | Affiliated Target |

|
| Class: Metabolism => Glycan biosynthesis and metabolism | Pathway Hierarchy | ||
| Glycosaminoglycan degradation | hsa00531 | Affiliated Target |

|
| Class: Metabolism => Glycan biosynthesis and metabolism | Pathway Hierarchy | ||
| Sphingolipid metabolism | hsa00600 | Affiliated Target |

|
| Class: Metabolism => Lipid metabolism | Pathway Hierarchy | ||
| Glycosphingolipid biosynthesis - ganglio series | hsa00604 | Affiliated Target |

|
| Class: Metabolism => Glycan biosynthesis and metabolism | Pathway Hierarchy | ||
| Lysosome | hsa04142 | Affiliated Target |

|
| Class: Cellular Processes => Transport and catabolism | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 8 | Degree centrality | 8.59E-04 | Betweenness centrality | 1.22E-03 |
|---|---|---|---|---|---|
| Closeness centrality | 1.37E-01 | Radiality | 1.14E+01 | Clustering coefficient | 1.79E-01 |
| Neighborhood connectivity | 4.13E+00 | Topological coefficient | 2.15E-01 | Eccentricity | 13 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Turnover of beta-galactosidase in fibroblasts from patients with genetically different types of beta-galactosidase deficiency. Biochem J. 1981 Oct 15;200(1):143-51. | |||||
| REF 2 | Clinical pipeline report, company report or official report of Axovant Gene Therapies | |||||
| REF 3 | ClinicalTrials.gov (NCT04273269) An Open-Label Adaptive-Design Study of Intracisternal Adenoassociated Viral Vector Serotype rh.10 Carrying the Human beta-Galactosidase cDNA for Treatment of GM1 Gangliosidosis. U.S.National Institutes of Health. | |||||
| REF 4 | ClinicalTrials.gov (NCT04713475) Phase 1/2 Open-Label, Multicenter Study to Assess the Safety, Tolerability and Efficacy of a Single Dose of PBGM01 Delivered Into the Cisterna Magna of Pediatric Type 1 (Early Onset) and Type 2a (Late Onset) Infantile GM1 Gangliosidosis. U.S.National Institutes of Health. | |||||
| REF 5 | Lysosomes as a therapeutic target. Nat Rev Drug Discov. 2019 Dec;18(12):923-948. | |||||
| REF 6 | Clinical pipeline report, company report or official report of Passage Bio | |||||
| REF 7 | A bicyclic 1-deoxygalactonojirimycin derivative as a novel pharmacological chaperone for GM1 gangliosidosis. Mol Ther. 2013 Mar;21(3):526-32. | |||||
| REF 8 | Chemical modification of the beta-glucocerebrosidase inhibitor N-octyl-beta-valienamine: synthesis and biological evaluation of 4-epimeric and 4-O-(beta-D-galactopyranosyl) derivatives. Bioorg Med Chem. 2002 Jun;10(6):1967-72. | |||||
| REF 9 | Crystal structure of human beta-galactosidase: structural basis of Gm1 gangliosidosis and morquio B diseases. J Biol Chem. 2012 Jan 13;287(3):1801-12. | |||||
| REF 10 | Structural basis of pharmacological chaperoning for human beta-galactosidase | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

