Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T72835
(Former ID: TTDC00098)
|
|||||
| Target Name |
E-selectin (SELE)
|
|||||
| Synonyms |
Leukocyte-endothelial cell adhesion molecule 2; LECAM2; Endothelial leukocyte adhesion molecule 1; ELAM1; ELAM-1; CD62E antigen; CD62E; CD62 antigen-like family member E
Click to Show/Hide
|
|||||
| Gene Name |
SELE
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 7 Target-related Diseases | + | ||||
| 1 | Acute myeloid leukaemia [ICD-11: 2A60] | |||||
| 2 | Asthma [ICD-11: CA23] | |||||
| 3 | Circulatory system disease [ICD-11: BE2Z] | |||||
| 4 | Hypertension [ICD-11: BA00-BA04] | |||||
| 5 | Atopic eczema [ICD-11: EA80] | |||||
| 6 | Chronic obstructive pulmonary disease [ICD-11: CA22] | |||||
| 7 | Psoriasis [ICD-11: EA90] | |||||
| Function |
Mediates in the adhesion of blood neutrophils in cytokine-activated endothelium through interaction with SELPLG/PSGL1. May have a role in capillary morphogenesis. Cell-surface glycoprotein having a role in immunoadhesion.
Click to Show/Hide
|
|||||
| UniProt ID | ||||||
| Sequence |
MIASQFLSALTLVLLIKESGAWSYNTSTEAMTYDEASAYCQQRYTHLVAIQNKEEIEYLN
SILSYSPSYYWIGIRKVNNVWVWVGTQKPLTEEAKNWAPGEPNNRQKDEDCVEIYIKREK DVGMWNDERCSKKKLALCYTAACTNTSCSGHGECVETINNYTCKCDPGFSGLKCEQIVNC TALESPEHGSLVCSHPLGNFSYNSSCSISCDRGYLPSSMETMQCMSSGEWSAPIPACNVV ECDAVTNPANGFVECFQNPGSFPWNTTCTFDCEEGFELMGAQSLQCTSSGNWDNEKPTCK AVTCRAVRQPQNGSVRCSHSPAGEFTFKSSCNFTCEEGFMLQGPAQVECTTQGQWTQQIP VCEAFQCTALSNPERGYMNCLPSASGSFRYGSSCEFSCEQGFVLKGSKRLQCGPTGEWDN EKPTCEAVRCDAVHQPPKGLVRCAHSPIGEFTYKSSCAFSCEEGFELHGSTQLECTSQGQ WTEEVPSCQVVKCSSLAVPGKINMSCSGEPVFGTVCKFACPEGWTLNGSAARTCGATGHW SGLLPTCEAPTESNIPLVAGLSAAGLSLLTLAPFLLWLRKCLRKAKKFVPASSCQSLESD GSYQKPSYIL Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| HIT2.0 ID | T24U57 | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 6 Clinical Trial Drugs | + | ||||
| 1 | GMI-1070 | Drug Info | Phase 3 | Asthma | [1], [2], [3] | |
| 2 | GMI-1271 | Drug Info | Phase 3 | Acute myeloid leukaemia | [4] | |
| 3 | CY-1503 | Drug Info | Phase 2/3 | Hypertension | [5] | |
| 4 | Bimosiamose | Drug Info | Phase 2a | Atopic dermatitis | [6] | |
| 5 | GMI-1359 | Drug Info | Phase 1 | Solid tumour/cancer | [7] | |
| 6 | PF-07209326 | Drug Info | Phase 1 | Sickle-cell disorder | [8] | |
| Discontinued Drug(s) | [+] 2 Discontinued Drugs | + | ||||
| 1 | SMART anti-E/P selectin | Drug Info | Discontinued in Phase 1 | Asthma | [9] | |
| 2 | GI-270384X | Drug Info | Terminated | Inflammatory bowel disease | [10] | |
| Mode of Action | [+] 3 Modes of Action | + | ||||
| Inhibitor | [+] 7 Inhibitor drugs | + | ||||
| 1 | GMI-1070 | Drug Info | [1], [3], [11] | |||
| 2 | GMI-1271 | Drug Info | [12] | |||
| 3 | Bimosiamose | Drug Info | [14], [15], [16] | |||
| 4 | 1na | Drug Info | [19] | |||
| 5 | Efomycine M | Drug Info | [20] | |||
| 6 | Fucose | Drug Info | [19] | |||
| 7 | O-Sialic Acid | Drug Info | [21] | |||
| Modulator | [+] 3 Modulator drugs | + | ||||
| 1 | CY-1503 | Drug Info | [13] | |||
| 2 | SMART anti-E/P selectin | Drug Info | [18] | |||
| 3 | GI-270384X | Drug Info | [10] | |||
| Antagonist | [+] 2 Antagonist drugs | + | ||||
| 1 | GMI-1359 | Drug Info | [17] | |||
| 2 | PF-07209326 | Drug Info | [8] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: (1R,2R,3S)-3-methylcyclohexane-1,2-diol | Ligand Info | |||||
| Structure Description | E-selectin lectin, EGF-like and two SCR domains complexed with glycomimetic antagonist | PDB:4C16 | ||||
| Method | X-ray diffraction | Resolution | 1.93 Å | Mutation | No | [22] |
| PDB Sequence |
WSYNTSTEAM
10 TYDEASAYCQ20 QRYTHLVAIQ30 NKEEIEYLNS40 ILSYSPSYYW50 IGIRKVNNVW 60 VWVGTQKPLT70 EEAKNWAPGE80 PNNRQKDEDC90 VEIYIKREKD100 VGMWNDERCS 110 KKKLALCYTA120 ACTNTSCSGH130 GECVETINNY140 TCKCDPGFSG150 LKCEQIVNCT 160 ALESPEHGSL170 VCSHPLGNFS180 YNSSCSISCD190 RGYLPSSMET200 MQCMSSGEWS 210 APIPACNVVE220 CDAVTNPANG230 FVECFQNPGS240 FPWNTTCTFD250 CEEGFELMGA 260 QSLQCTSSGN270 WDNEKPTCKA280
|
|||||
|
|
||||||
| Ligand Name: (S)-Cyclohexyl Lactic Acid | Ligand Info | |||||
| Structure Description | E-selectin lectin, EGF-like and two SCR domains complexed with glycomimetic antagonist | PDB:4C16 | ||||
| Method | X-ray diffraction | Resolution | 1.93 Å | Mutation | No | [22] |
| PDB Sequence |
WSYNTSTEAM
10 TYDEASAYCQ20 QRYTHLVAIQ30 NKEEIEYLNS40 ILSYSPSYYW50 IGIRKVNNVW 60 VWVGTQKPLT70 EEAKNWAPGE80 PNNRQKDEDC90 VEIYIKREKD100 VGMWNDERCS 110 KKKLALCYTA120 ACTNTSCSGH130 GECVETINNY140 TCKCDPGFSG150 LKCEQIVNCT 160 ALESPEHGSL170 VCSHPLGNFS180 YNSSCSISCD190 RGYLPSSMET200 MQCMSSGEWS 210 APIPACNVVE220 CDAVTNPANG230 FVECFQNPGS240 FPWNTTCTFD250 CEEGFELMGA 260 QSLQCTSSGN270 WDNEKPTCKA280
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Pathway Affiliation
Biological Network Descriptors
|
|
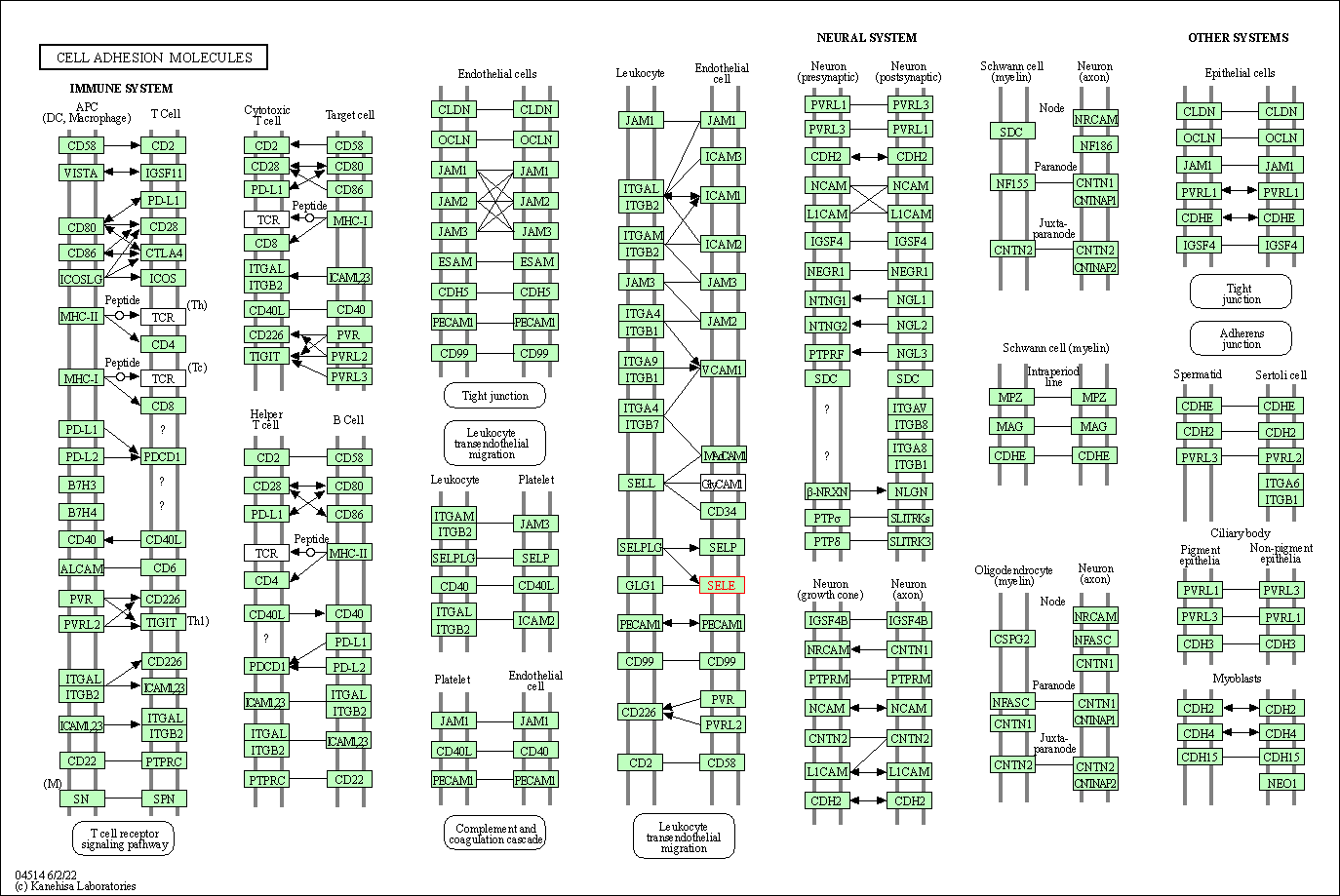
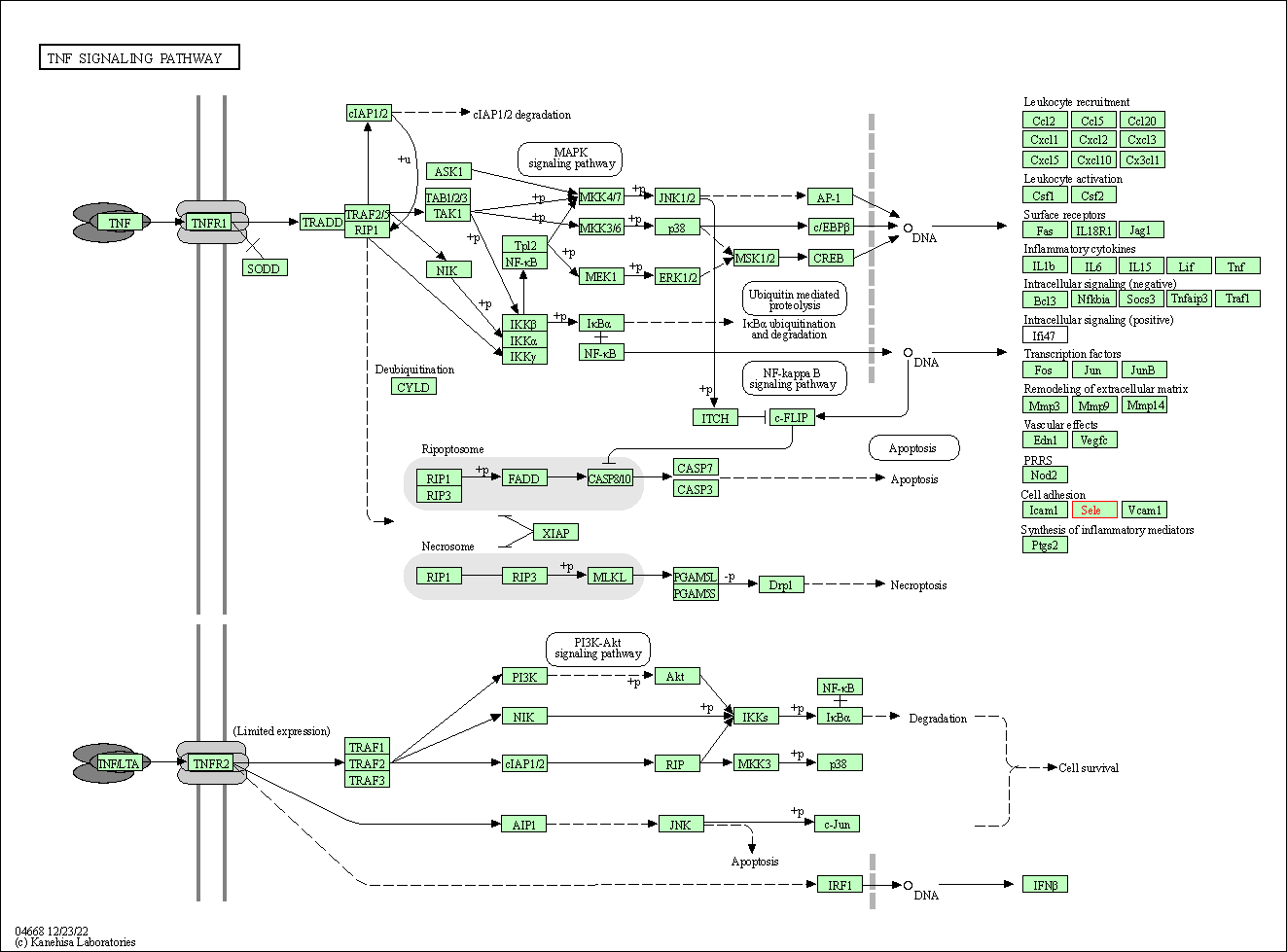
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Cell adhesion molecules | hsa04514 | Affiliated Target |

|
| Class: Environmental Information Processing => Signaling molecules and interaction | Pathway Hierarchy | ||
| TNF signaling pathway | hsa04668 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Degree | 5 | Degree centrality | 5.37E-04 | Betweenness centrality | 2.52E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 2.10E-01 | Radiality | 1.37E+01 | Clustering coefficient | 0.00E+00 |
| Neighborhood connectivity | 1.74E+01 | Topological coefficient | 2.22E-01 | Eccentricity | 13 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-regulating microRNAs | ||||||
| Target-regulating Transcription Factors | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) |
||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 4 KEGG Pathways | + | ||||
| 1 | Cell adhesion molecules (CAMs) | |||||
| 2 | TNF signaling pathway | |||||
| 3 | African trypanosomiasis | |||||
| 4 | Malaria | |||||
| NetPath Pathway | [+] 4 NetPath Pathways | + | ||||
| 1 | IL5 Signaling Pathway | |||||
| 2 | IL4 Signaling Pathway | |||||
| 3 | TNFalpha Signaling Pathway | |||||
| 4 | ID Signaling Pathway | |||||
| PID Pathway | [+] 3 PID Pathways | + | ||||
| 1 | Thromboxane A2 receptor signaling | |||||
| 2 | Glucocorticoid receptor regulatory network | |||||
| 3 | ATF-2 transcription factor network | |||||
| Reactome | [+] 1 Reactome Pathways | + | ||||
| 1 | Cell surface interactions at the vascular wall | |||||
| WikiPathways | [+] 3 WikiPathways | + | ||||
| 1 | Human Complement System | |||||
| 2 | TNF alpha Signaling Pathway | |||||
| 3 | Cell surface interactions at the vascular wall | |||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Deal watch: Pfizer deal for selectin inhibitor highlights potential of glycomimetic drugs. Nat Rev Drug Discov. 2011 Dec 1;10(12):890. | |||||
| REF 2 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 8307). | |||||
| REF 3 | GMI-1070, a novel pan-selectin antagonist, reverses acute vascular occlusions in sickle cell mice. Blood. 2010 Sep 9;116(10):1779-86. | |||||
| REF 4 | ClinicalTrials.gov (NCT03616470) Study to Determine the Efficacy of Uproleselan (GMI-1271) in Combination With Chemotherapy to Treat Relapsed/Refractory Acute Myeloid Leukemia. U.S. National Institutes of Health. | |||||
| REF 5 | ClinicalTrials.gov (NCT00226369) Cylexin for Reduction of Reperfusion Injury in Infant Heart Surgery. U.S. National Institutes of Health. | |||||
| REF 6 | Emerging drugs for asthma. Expert Opin Emerg Drugs. 2008 Dec;13(4):643-53. | |||||
| REF 7 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 8 | ClinicalTrials.gov (NCT04255875) A RANDOMIZED, DOUBLE-BLIND, PLACEBO CONTROLLED EVALUATION OF SINGLE DOSES OF PF-07209326 IN HEALTHY PARTICIPANTS (SAFETY, TOLERABILITY, AND PHARMACOKINETICS [PK]) FOLLOWED BY AN OPEN LABEL, REPEAT DOSE EVALUATION IN SICKLE CELL DISEASE PARTICIPANTS (SAFETY, TOLERABILITY, PK AND EFFICACY). U.S.National Institutes of Health. | |||||
| REF 9 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800008129) | |||||
| REF 10 | Inhibition of endothelial cell adhesion molecule expression improves colonic hyperalgaesia. Neurogastroenterol Motil. 2009 Feb;21(2):189-96. | |||||
| REF 11 | GMI-1070, a novel pan-selectin antagonist, reverses acute vascular occlusions in sickle cell mice. Blood. 2010 September 9; 116(10): 1779-1786. | |||||
| REF 12 | Clinical pipeline report, company report or official report of glycomimetics. | |||||
| REF 13 | Adjunctive selectin blockade successfully reduces infarct size beyond thrombolysis in the electrolytic canine coronary artery model. Circulation. 1995 Aug 1;92(3):492-9. | |||||
| REF 14 | Effects of the pan-selectin antagonist bimosiamose (TBC1269) in experimental human endotoxemia. Shock. 2008 Apr;29(4):475-82. | |||||
| REF 15 | The pharmacokinetics of subcutaneously injected Bimosiamose disodium in healthy male volunteers. Biopharm Drug Dispos. 2007 Dec;28(9):475-84. | |||||
| REF 16 | Bimosiamose, an inhaled small-molecule pan-selectin antagonist, attenuates late asthmatic reactions following allergen challenge in mild asthmatics... Pulm Pharmacol Ther. 2006;19(4):233-41. | |||||
| REF 17 | Clinical pipeline report, company report or official report of GlycoMimetics. | |||||
| REF 18 | HuEP5C7 as a humanized monoclonal anti-E/P-selectin neurovascular protective strategy in a blinded placebo-controlled trial of nonhuman primate stroke. Circ Res. 2002 Nov 15;91(10):907-14. | |||||
| REF 19 | How many drug targets are there Nat Rev Drug Discov. 2006 Dec;5(12):993-6. | |||||
| REF 20 | Interfering with leukocyte rolling--a promising therapeutic approach in inflammatory skin disorders Trends Pharmacol Sci. 2003 Feb;24(2):49-52. | |||||
| REF 21 | The Protein Data Bank. Nucleic Acids Res. 2000 Jan 1;28(1):235-42. | |||||
| REF 22 | E-selectin ligand complexes adopt an extended high-affinity conformation. J Mol Cell Biol. 2016 Feb;8(1):62-72. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

