Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T68877
(Former ID: TTDR00810)
|
|||||
| Target Name |
DNA-binding protein inhibitor ID-2 (ID2)
|
|||||
| Synonyms |
bHLHb26; Inhibitor of differentiation 2; Inhibitor of DNA binding 2; Id2; Class B basic helix-loop-helix protein 26
Click to Show/Hide
|
|||||
| Gene Name |
ID2
|
|||||
| Target Type |
Literature-reported target
|
[1] | ||||
| Function |
Implicated in regulating a variety of cellular processes, including cellular growth, senescence, differentiation, apoptosis, angiogenesis, and neoplastic transformation. Inhibits skeletal muscle and cardiac myocyte differentiation. Regulates the circadian clock by repressing the transcriptional activator activity of the CLOCK-ARNTL/BMAL1 heterodimer. Restricts the CLOCK and ARNTL/BMAL1 localization to the cytoplasm. Plays a role in both the input and output pathways of the circadian clock: in the input component, is involved in modulating the magnitude of photic entrainment and in the output component, contributes to the regulation of a variety of liver clock-controlled genes involved in lipid metabolism. Transcriptional regulator (lacking a basic DNA binding domain) which negatively regulates the basic helix-loop-helix (bHLH) transcription factors by forming heterodimers and inhibiting their DNA binding and transcriptional activity.
Click to Show/Hide
|
|||||
| UniProt ID | ||||||
| Sequence |
MKAFSPVRSVRKNSLSDHSLGISRSKTPVDDPMSLLYNMNDCYSKLKELVPSIPQNKKVS
KMEILQHVIDYILDLQIALDSHPTIVSLHHQRPGQNQASRTPLTTLNTDISILSLQASEF PSELMSNDSKALCG Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Pathway Affiliation
Biological Network Descriptors
|
|
| Protein Name | Pfam ID | Percentage of Identity (%) | E value |
|---|---|---|---|
| Putative DNA-binding protein inhibitor ID-2B (ID2B) | 86.111 (31/36) | 8.70E-15 | |
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| TGF-beta signaling pathway | hsa04350 | Affiliated Target |
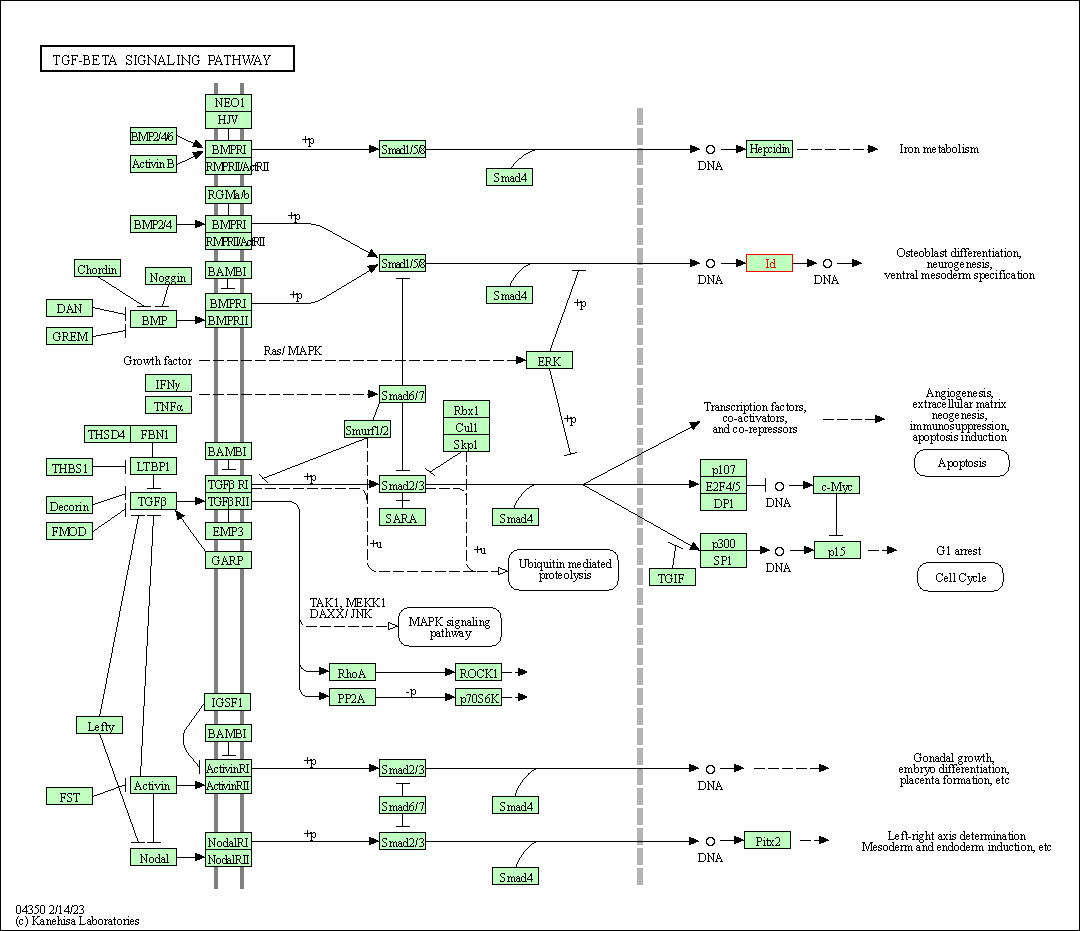
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Hippo signaling pathway | hsa04390 | Affiliated Target |
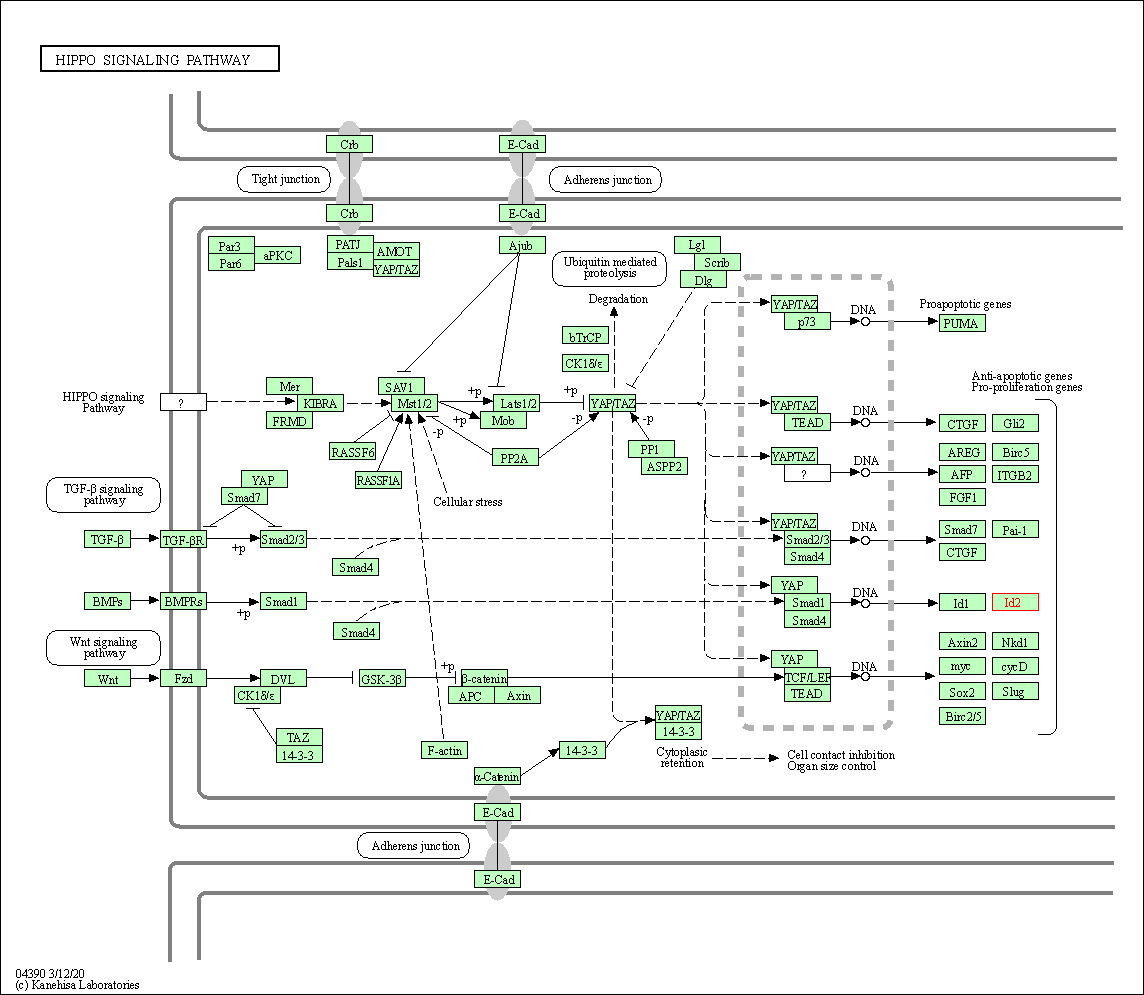
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Signaling pathways regulating pluripotency of stem cells | hsa04550 | Affiliated Target |
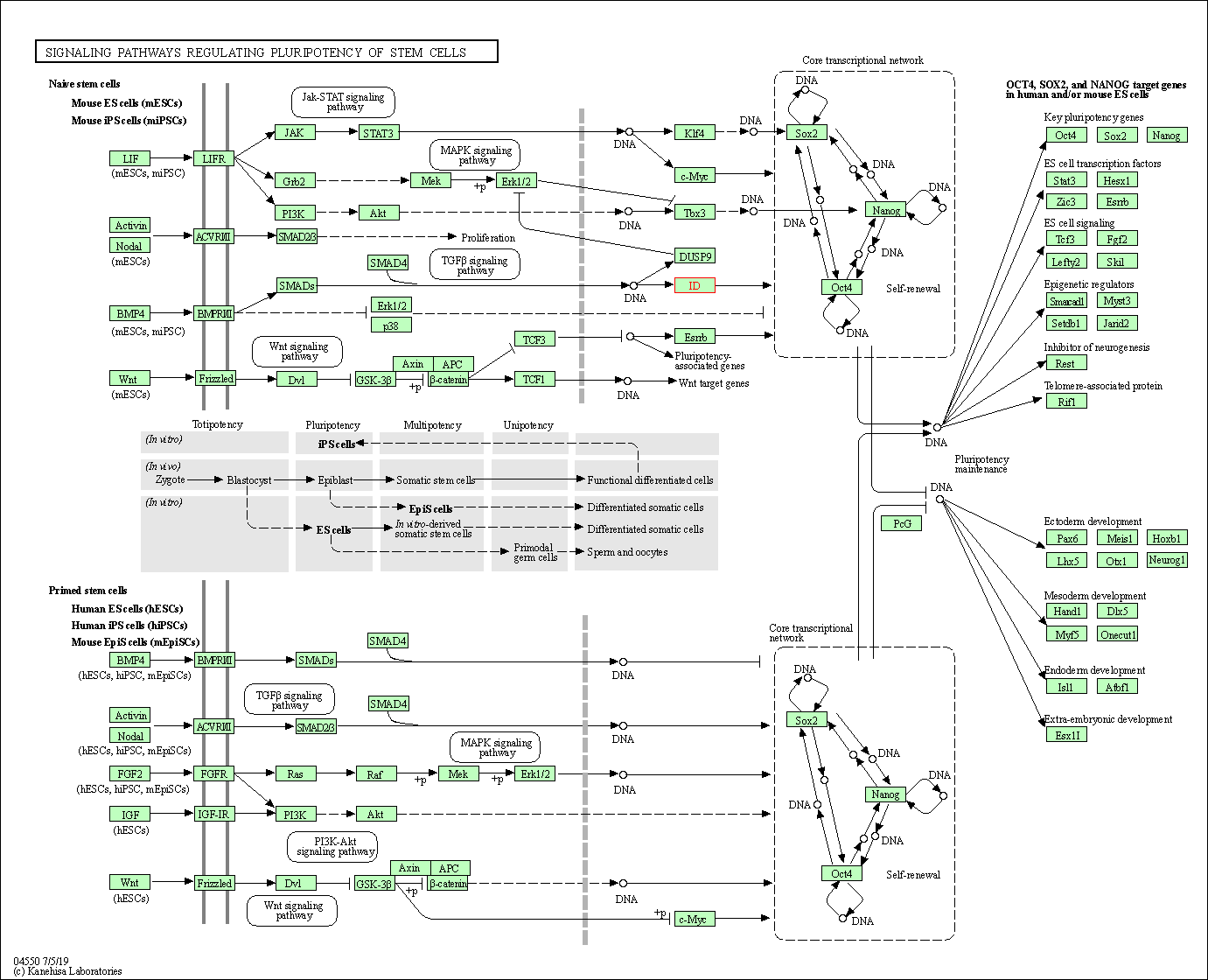
|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
| Degree | 6 | Degree centrality | 6.45E-04 | Betweenness centrality | 5.05E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 2.10E-01 | Radiality | 1.37E+01 | Clustering coefficient | 6.67E-02 |
| Neighborhood connectivity | 1.53E+01 | Topological coefficient | 1.88E-01 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-interacting Proteins | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | TH2 dominance and defective development of a CD8+ dendritic cell subset in Id2-deficient mice. J Allergy Clin Immunol. 2003 Jan;111(1):136-42. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

